Elevated NR2F1 underlies the persistence of invasive disease after treatment of BRAF-mutant melanoma
- PMID: 40955663
- PMCID: PMC12435839
- DOI: 10.1172/JCI178446
Elevated NR2F1 underlies the persistence of invasive disease after treatment of BRAF-mutant melanoma
Abstract
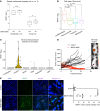
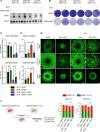
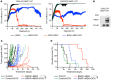
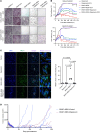
Despite the success of targeted inhibitors in cutaneous melanoma, therapeutic responses are limited by the aged tumor microenvironment and drug-tolerant residual cells. Given the similarities between drug tolerance and cellular dormancy, we studied the dormancy marker, nuclear receptor subfamily 2 group F member 1 (NR2F1), in response to BRAF-V600E inhibitors (BRAFi) plus MEK inhibitors (MEKi) in BRAF-mutant melanoma models. Transcriptomic analysis of melanoma patient samples treated with BRAFi + MEKi showed increased NR2F1. NR2F1 was highly expressed in the drug-tolerant invasive cell state of minimal residual disease in patient-derived and mouse-derived xenografts on BRAFi + MEKi. NR2F1 over-expression was sufficient to reduce BRAFi + MEKi effects on tumor growth in vivo, and cell proliferation, death, and invasion in vitro. Effects were linked to genes involved in mTORC1 signaling. These cells were sensitive to the combination of BRAFi, MEKi plus rapamycin. Melanomas from aged mice, known to exhibit decreased responses to BRAFi + MEKi, displayed higher levels of NR2F1 compared to tumors from young mice. Depleting NR2F1 in an aged mouse melanomas improved the response to targeted therapy. These findings show high NR2F1 expression in 'invasive-state' residual cells and that targeting NR2F1-high cells with mTORC1 inhibitors may improve outcomes in patients with melanoma.
Keywords: Drug therapy; Melanoma; Oncology; Therapeutics.
Conflict of interest statement
Figures


Comment in
- NR2F1 and mTORC1 provide the bridge between melanoma dormancy and therapeutic resistance doi: 10.1172/JCI197764
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

