Maintenance DNA methylation is required for induced Treg reparative function following viral pneumonia in mice
- PMID: 40956625
- PMCID: PMC12618063
- DOI: 10.1172/JCI192925
Maintenance DNA methylation is required for induced Treg reparative function following viral pneumonia in mice
Abstract
FOXP3+ natural regulatory T cells (nTregs) promote resolution of inflammation and repair of epithelial damage following viral pneumonia-induced lung injury, thus representing a cellular therapy for patients with severe viral pneumonia and the acute respiratory distress syndrome. Whether in vitro-induced Tregs (iTregs), which can be rapidly generated in substantial numbers from conventional T cells, also promote lung recovery is unknown. nTregs require specific DNA methylation patterns maintained by the epigenetic regulator ubiquitin-like with PHD and RING finger domains 1 (UHRF1). Here, we tested whether iTregs promote recovery following viral pneumonia and whether iTregs require UHRF1 for their pro-recovery function. We found that adoptive transfer of iTregs to mice with influenza virus pneumonia promotes lung recovery and that loss of UHRF1-mediated maintenance DNA methylation in iTregs leads to reduced engraftment and a delayed repair response. Transcriptional and DNA methylation profiling of adoptively transferred UHRF1-deficient iTregs that had trafficked to influenza-injured lungs demonstrated transcriptional instability with gain of transcription factors that define effector T cell lineage. Strategies to promote the stability of iTregs could be leveraged to further augment their pro-recovery function during viral pneumonia and other causes of severe lung injury.
Keywords: Epigenetics; Immunology; Inflammation; Influenza; Pulmonology; T cells.
Conflict of interest statement
Figures
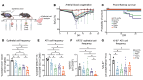


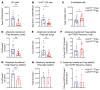

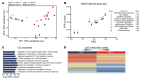
Update of
-
Maintenance DNA methylation is required for induced regulatory T cell reparative function following viral pneumonia.bioRxiv [Preprint]. 2025 Mar 1:2025.02.25.640199. doi: 10.1101/2025.02.25.640199. bioRxiv. 2025. Update in: J Clin Invest. 2025 Sep 16;135(22):e192925. doi: 10.1172/JCI192925. PMID: 40060513 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 HL153122/HL/NHLBI NIH HHS/United States
- U19 AI181102/AI/NIAID NIH HHS/United States
- K08 HL159356/HL/NHLBI NIH HHS/United States
- P01 AG049665/AG/NIA NIH HHS/United States
- U19 AI135964/AI/NIAID NIH HHS/United States
- P01 HL154998/HL/NHLBI NIH HHS/United States
- T32 AI083216/AI/NIAID NIH HHS/United States
- F31 HL162490/HL/NHLBI NIH HHS/United States
- R01 HL149883/HL/NHLBI NIH HHS/United States
- T32 GM144295/GM/NIGMS NIH HHS/United States
- F32 HL162418/HL/NHLBI NIH HHS/United States
- T32 HL076139/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

