Prophage regulation of Shewanella fidelis 3313 motility and biofilm formation with implications for gut colonization dynamics in Ciona robusta
- PMID: 40956701
- PMCID: PMC12440353
- DOI: 10.7554/eLife.103107
Prophage regulation of Shewanella fidelis 3313 motility and biofilm formation with implications for gut colonization dynamics in Ciona robusta
Abstract
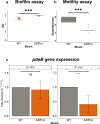
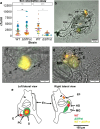
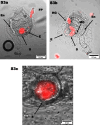
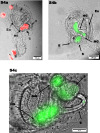
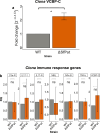
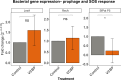
Lysogens, bacteria with one or more viruses (prophages) integrated into their genomes, are abundant in the gut of animals. Prophages often influence bacterial traits; however, the influence of prophages on the gut microbiota-host immune axis in animals remains poorly understood. Here, we investigate the influence of the prophage SfPat on Shewanella fidelis 3313, a persistent member of the gut microbiome of the model marine tunicate, Ciona robusta. Establishment of a SfPat deletion mutant (ΔSfPat) reveals the influence of this prophage on bacterial physiology in vitro and during colonization of the Ciona gut. In vitro, deletion of SfPat reduces S. fidelis 3313 motility and swimming while increasing biofilm formation. To understand the in vivo impact of these prophage-induced changes in bacterial traits, we exposed metamorphic stage 4 Ciona juveniles to wildtype (WT) and ΔSfPat strains. During colonization, ΔSfPat localizes to overlapping and distinct areas of the gut compared to the WT strain. We examined the differential expression of various regulators of cyclic-di-GMP, a secondary signaling molecule that mediates biofilm formation and motility. The pdeB gene, which encodes a bacterial phosphodiesterase known to influence biofilm formation and motility by degrading cyclic-di-GMP, is upregulated in the WT strain but not in ΔSfPat when examined in vivo. Expression of the Ciona gut immune effector, VCBP-C, is enhanced during colonization by ΔSfPat compared to the WT strain; however, VCBP-C binding to the WT strain does not promote the excision of SfPat in an SOS-dependent pathway. Instead, VCBP-C binding significantly reduces the expression of a phage major capsid protein. Our findings suggest that SfPat influences host perception of this important colonizing commensal and highlights the significance of investigating tripartite dynamics between prophages, bacteria, and their animal hosts to better understand the gut microbiota-host immune axis.
Keywords: Ciona robusta; Shewanella fidelis 3313; VCBP-C; ciona robusta (c. intestinalis type a); gut microbiome; infectious disease; innate immunity; microbiology; prophage; tunicate.
© 2025, Natarajan et al.
Conflict of interest statement
ON, SG, MY, SL, FN, NP, CA, AL, EK, BL, MB, JG, LD No competing interests declared
Figures




Update of
References
-
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. - DOI - PubMed
-
- Aron EJ, Trimble The bee swarm plot, an alternative to stripchart. d641db5Github. 2021 https://github.com/aroneklund/beeswarm
-
- Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. Journal of Bacteriology. 2005;187:3311–3318. doi: 10.1128/JB.187.10.3311-3318.2005. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

