AI-Validated Brain Targeted mRNA Lipid Nanoparticles with Neuronal Tropism
- PMID: 40957853
- PMCID: PMC12548354
- DOI: 10.1021/acsnano.4c15013
AI-Validated Brain Targeted mRNA Lipid Nanoparticles with Neuronal Tropism
Abstract
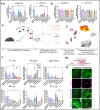
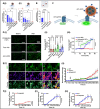
Targeting therapeutic nanoparticles to the brain poses a challenge due to the restrictive nature of the blood-brain barrier (BBB). Here we report the development of mRNA-loaded lipid nanoparticles (LNPs) functionalized with BBB-interacting small molecules, thereby enhancing brain delivery and gene expression. Screening brain-targeted mRNA-LNPs in central nervous system (CNS) in vitro models and through intravenous administration in mice demonstrated that acetylcholine-conjugated LNPs achieved superior brain tropism and gene expression, outperforming LNP modifications with nicotine, glucose, memantine, cocaine, tryptophan, and other small molecules. An artificial intelligence (AI)-based model designed to predict the BBB permeability of small-molecule ligands showed strong alignment with our experimental results, providing in vivo validation of its predictive capacity. Cell-specific biodistribution analysis in Cre-reporter Ai9 mice showed that acetylcholine-functionalized LNPs preferentially transfected neurons and astrocytes following either intravenous or intracerebral administration. Mechanistic studies suggest that acetylcholine-LNP uptake is mediated by the functional engagement of acetylcholine receptors (AchRs) followed by endocytosis, which synergistically enhances intracellular mRNA delivery. Moreover, acetylcholine-LNPs successfully crossed a human BBB-on-a-chip model, enabling transgene expression in human iPSC-derived neurons. Their effective penetration and transfection in human brain organoids further support their potential activity in human-based systems. These findings establish a predictive and modular framework for engineering CNS-targeted LNPs, advancing precision gene delivery for brain disorders.
Keywords: Artificial Intelligence; Blood−Brain Barrier; Brain Targeting; Central Nervous System (CNS); Gene Delivery; Lipid Nanoparticles; mRNA.
Figures




References
-
- Li H., Cao Y., Ye J., Yang Z., Chen Q., Liu X., Zhang B., Qiao J., Tang Q., Yang H., Li J., Shi Z., Mao Y.. Engineering brain-derived neurotrophic factor mRNA delivery for the treatment of Alzheimer’s disease. Chem. Eng. J. 2023;466:143152. doi: 10.1016/j.cej.2023.143152. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

