Transgenerational inheritance of diminished ovarian reserve triggered by prenatal propylparaben exposure in mice
- PMID: 40957879
- PMCID: PMC12441155
- DOI: 10.1038/s41467-025-63440-z
Transgenerational inheritance of diminished ovarian reserve triggered by prenatal propylparaben exposure in mice
Abstract
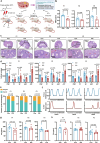
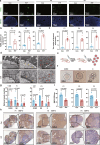
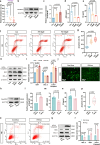
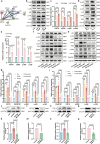
Diminished ovarian reserve (DOR) is associated with heightened risk of infertility, premature menopause, and various long-term health issues. Our previous research demonstrated a correlation between prenatal propylparaben exposure and DOR in F1 mice. Here, we further reveal that the DOR phenotypes can be transgenerationally inherited in F1-F3 mice, manifested through increased follicular atresia and decreased anti-Müllerian hormone levels. Excessive apoptosis of granulosa cells is found to underlie these pathological processes. By combining diverse sequencing techniques, we identify persistent Rhobtb1 hypomethylation across multiple generations. Further exploration reveals that RhoBTB1 regulates FGF18 via ubiquitination, triggering MAPK pathway activation and subsequent granulosa cell apoptosis. Notably, similar Rhobtb1 hypomethylation patterns are observed in blood samples from DOR patients. Furthermore, intervention with a methyl-donor diet effectively ameliorates DOR phenotypes in F1-F3 offspring. These findings highlight the transgenerational effects of DOR, elucidate its underlying causes and pathogenic mechanisms, and propose potential epigenetic therapy strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Sharara, F. I., Scott, R. T. Jr. & Seifer, D. B. The detection of diminished ovarian reserve in infertile women. Am. J. Obstet. Gynecol.179, 804–812 (1998). - PubMed
-
- Barker, D. J. & Osmond, C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet1, 1077–1081 (1986). - PubMed
MeSH terms
Substances
Grants and funding
- 82371648/National Natural Science Foundation of China (National Science Foundation of China)
- 82471678/National Natural Science Foundation of China (National Science Foundation of China)
- 2025AFB052/Natural Science Foundation of Hubei Province (Hubei Provincial Natural Science Foundation)
- 82101703/National Science Foundation of China | Young Scientists Fund
- 2021YFC2700402/National Science Foundation of China | Key Programme
LinkOut - more resources
Full Text Sources
Miscellaneous

