A mesothelial differentiation gateway drives fibrosis
- PMID: 40957887
- PMCID: PMC12441157
- DOI: 10.1038/s41467-025-63990-2
A mesothelial differentiation gateway drives fibrosis
Abstract
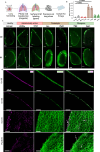
Internal organs are encased by a supportive epithelial monolayer of mesodermal origin, termed mesothelium. The nature, evolution and function of mesothelial cells, and their genetic regulation impacting disease development are insufficiently understood. Here, we generate a comprehensive organ-wide single-cell transcriptomic compendium of mesothelium across healthy and diseased mouse and human organs, delineating the evolution of conserved activated states of mesothelial cells in response to disease. We uncover genetic drives behind each cell state and reveal a conserved metabolic gate into multipotent proteolytic, inflammatory and fibrotic cell differentiation, in mouse and human. Using lung injury models in mice, in combination with mesothelial cell-specific viral approaches, we show that direct metabolic reprogramming using Ifi27l2a and Crip1 on organ surfaces, blocks multipotent differentiation and protects mouse lungs from fibrotic disease. These findings place mesothelial cells as cellular exemplars and gateway to fibrotic disease, opening translational approaches to subvert fibrosis across a range of clinical indications.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: A.F., M.M., S.K., and Y.R. have filed patent application WO2023079127A1 covering the use of these methods to study and manipulate mesothelial cells in lung fibrosis. The remaining authors declare no competing interests.
Figures






References
-
- Wang, N. S. The regional difference of pleural mesothelial cells in rabbits. Am. Rev. Respir. Dis.110, 623–633 (1974). - PubMed
-
- Mutsaers, S. E. & Wilkosz, S. Structure and function of mesothelial cells. Cancer Treat. Res.134, 1–19 (2007). - PubMed
-
- Mutsaers, S. E., Prêle, C. M.-A., Pengelly, S. & Herrick, S. E. Mesothelial cells and peritoneal homeostasis. Fertil. Steril.106, 1018–1024 (2016). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

