Transcriptional dynamics of the oligodendrocyte lineage and its regulation by the brain erythropoietin system
- PMID: 40957896
- PMCID: PMC12441126
- DOI: 10.1038/s41467-025-62791-x
Transcriptional dynamics of the oligodendrocyte lineage and its regulation by the brain erythropoietin system
Abstract
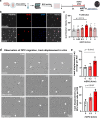
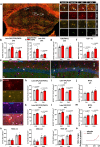
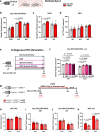
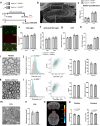
Oligodendrocytes differentiate from oligodendrocyte progenitor cells (OPC) in early postnatal development, but some oligodendrogenesis is maintained throughout adulthood, where oligodendrocyte lineage dynamics may contribute to neuroplasticity, adaptive myelination, and myelin repair. Here, we studied the effect of erythropoietin (EPO) and its receptor (EPOR) on oligodendrocyte lineage dynamics employing murine hippocampus and its myelinated fibers as model region. Using multiple stage-specific markers and single-nuclei-RNA-seq data, we find that EPO stimulates all oligodendroglial lineage cells directly, driving differentiation/maturation. Differential gene expression analysis reveals multiple EPO-regulated mRNAs, including downregulated transcripts for GABA-A receptors, fitting the known inhibition of oligodendrocyte maturation by GABA. Importantly, analogous oligodendrocyte responses are seen when endogenous EPO expression in brain is stimulated by hypoxia. Mice lacking EPOR from mature oligodendrocytes show subtle deficiencies of adult myelination in hippocampal fimbria and mild working memory deficits. These gain- and loss-of-function experiments may further suggest EPO as clinically safe treatment for remyelination therapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing financial or other interests in connection with this article.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

