SARS-CoV-2 infection dynamics in a MHCI-mismatched lung transplant recipient
- PMID: 40957936
- PMCID: PMC12441153
- DOI: 10.1038/s41467-025-63681-y
SARS-CoV-2 infection dynamics in a MHCI-mismatched lung transplant recipient
Abstract
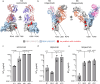
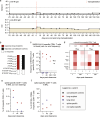
A 48-year-old patient underwent lung transplantation because of severe COVID-19, which aggravated his underlying interstitial lung disease, despite the presence of detectable SARS-CoV-2. Subsequently, the graft is re-infected early in the post-procedural phase, leading to viral persistence for more than five months. By analyzing viral evolution and effector immune response within the transplanted organ, we observe three main findings. First, virus evolution differs in the transplanted organ compared to that in the upper respiratory tract and is affected by monoclonal SARS-CoV-2-specific antibodies and molnupiravir. Second, we show the potential clinical relevance of T cell HLA restriction that may facilitate viral clearance in the upper respiratory tract compared to the ongoing viral replication in the HLA mismatch organ. Third, close monitoring and modulation of immunosuppressive and antiviral therapy enables viral clearance in a lung transplantation setting despite incomplete SARS-CoV-2 clearance prior to transplantation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.L. indicates research grants from the German Research Foundation, the German Heart Foundation and the German Center for Infectious Research. A.L. indicates travel grants from Diaplan and participation in advisory boards of Bayer AG. B.C.F. indicates research support from Bristol-Myer Squibb and Relief Therapeutics unrelated to the manuscript, consulting and lecture fees from Advita Lifescience GmbH, Actelion, AstraZeneca, Boehringer Ingelheim, Novartis, Roche and Vifor, travel support from Boehringer Ingelheim. B.C.F. indicates the following intellectual property: WO2020225246A1; WO2021152119A1. DH indicates lecture fees from BioMerieux. D.St. reports financial support for lectures or participation at advisory boards from Astra-Zeneca AG, Novartis AG, GSK AG, Roche AG, Zambon, Pfizer, Schwabe Pharma AG, Vifor AG, Chiesi AG, MSD, Pfizer, Sanofi, Chemie Menarini, CSL-Behring, Boehringer Ingelheim outside the submitted work. N.K. indicates lecture fees from AstraZeneca. PA indicates personal lecture fees from Boehringer Ingelheim. SF indicates personal lecture fees from Astra Zeneca and CSL Behring outside the submitted work. All other authors declare no competing interests.
Figures


References
-
- Bellani, G. et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. Jama315, 788–800 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

