Senescent Tumoral HLA-E Reshapes Microenvironment through Impairing NK Cell-Dendritic Cell-T Cell Network in Malignant Pleural Effusion from Lung Cancer
- PMID: 40959273
- PMCID: PMC12435339
- DOI: 10.7150/ijbs.116499
Senescent Tumoral HLA-E Reshapes Microenvironment through Impairing NK Cell-Dendritic Cell-T Cell Network in Malignant Pleural Effusion from Lung Cancer
Abstract
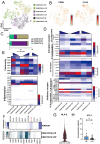
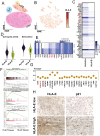
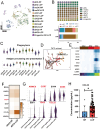
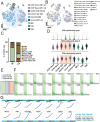
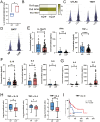
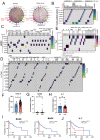
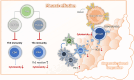
Background: Malignant pleural effusion (MPE) is ominous in lung cancer patients. However, comprehensive studies of both innate and adaptive immune responses within the pleural tumor microenvironment remain limited. Methods: We collected samples from patients with heart failure and lung cancer-MPE. By single-cell RNA sequencing, we analyzed alternations in cancer cells, NK cells, DCs, and T cells. Key cytokines involving in cell-cell interactions were quantified using Luminex or ELISA, while HLA-E and aging markers were assessed via immunohistochemistry. Results: Our findings revealed that CD56⁺CD16⁺ and CD56⁻CD16⁻ NK cells exhibited reduced cytotoxicity, mainly through HLA-E-expressing senescent cancer cells interacting with NK cells inhibitory receptor, leading to NK cell dysfunction and reduced XCL2 expression, which might impair cDC1 recruitment. Consequently, aDC2 cells evolved into exhausted phenotype, resulting in inadequate T cell activation. In CD8 T cells, transcription factors such as FOXO1 contributed to diminished cytotoxicity. Despite presence of GZMA CD4 T cells, their cytotoxicity was suppressed in MPE. Th1-like and Th2-like regulatory T cells further inhibited CD4 T cell responses. Key molecules, CXCL16, BAG6, and IL-7, bridging innate and adaptive immunity conferred poor prognosis. Conclusions: Our study demonstrates that senescent cancer cells promote immunoevasion through HLA-E, suppressing NK cell cytotoxicity, impairing DC function, and disrupting T cell activation. Cell-cell interaction and imbalanced Th1/Th2 contribute to microenvironmental remodeling, driving disease progression. These findings provide insights into the immunological landscape and therapeutic targets for intervention.
Keywords: HLA-E; dendritic cell; nature killer cell; pleural metastasis; senescent cancer cell.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Sahn SA. Pleural diseases related to metastatic malignancies. Eur Respir J. 1997;10:1907–13. - PubMed
-
- Yano S, Shinohara H, Herbst RS, Kuniyasu H, Bucana CD, Ellis LM. et al. Production of experimental malignant pleural effusions is dependent on invasion of the pleura and expression of vascular endothelial growth factor/vascular permeability factor by human lung cancer cells. Am J Pathol. 2000;157:1893–903. - PMC - PubMed
-
- Lee YC, Light RW. Management of malignant pleural effusions. Respirology. 2004;9:148–56. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

