Automated, high-throughput in situ hybridization of sea urchin (Lytechinus pictus) embryos
- PMID: 40960013
- PMCID: PMC12516323
- DOI: 10.1242/dev.204814
Automated, high-throughput in situ hybridization of sea urchin (Lytechinus pictus) embryos
Abstract
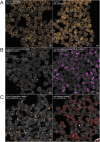
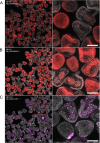
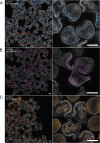
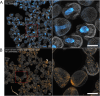
Despite the reach of in situ hybridization (ISH) in developmental biology, it is rarely used at scale. The major bottleneck is the throughput of the assay, which relies upon labor-intensive manual steps. The goal of this study was to develop a high-throughput, automated hybridization chain reaction (HCR) pipeline for the sea urchin (Lytechinus pictus). Our method, which we term high-throughput (HT)-HCR, can process 192 gene probe sets on whole-mount embryos within 32 h. The physical properties of sea urchin embryos enabled us to utilize a 96-well plate format, miniaturized reaction volumes, a general-purpose robotic liquid handler and automated confocal microscopy. Using this approach, we produced high quality localization data for 101 target genes across three developmental stages. The results reveal the localization of previously undescribed physiological genes, as well as canonical developmental transcription factors. HT-HCR represents an order of magnitude increase in the throughput of spatial expression profiling studies utilizing the sea urchin. This will enable more-sophisticated perturbation analyses and drug-screening efforts in this emerging animal model.
Keywords: In situ hybridization (ISH); Lytechinus pictus; High-throughput; Hybridization chain reaction (HCR); Sea urchin; Spatial transcriptomics; Transcription factor; Transporter.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



Update of
-
Automated, high-throughput in-situ hybridization of Lytechinus pictus embryos.bioRxiv [Preprint]. 2025 Mar 24:2025.03.23.644641. doi: 10.1101/2025.03.23.644641. bioRxiv. 2025. Update in: Development. 2025 Sep 15;152(18):dev204814. doi: 10.1242/dev.204814. PMID: 40196544 Free PMC article. Updated. Preprint.
References
-
- Anderson, S. L., Hose, J. E. and Knezovich, J. P. (1994). Genotoxic and developmental effects in sea urchins are sensitive indicators of effects of genotoxic chemicals. Environ. Toxicol. Chem. 13, 1033-1041. 10.1002/etc.5620130704 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

