Mapping the genetic landscape of iron metabolism uncovers the SETD2 methyltransferase as a modulator of iron flux
- PMID: 40961184
- PMCID: PMC12442849
- DOI: 10.1126/sciadv.adw9095
Mapping the genetic landscape of iron metabolism uncovers the SETD2 methyltransferase as a modulator of iron flux
Abstract
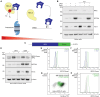
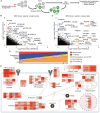
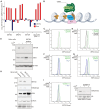
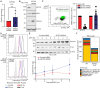
Cellular iron levels must be tightly regulated to ensure sufficient iron for essential enzymatic functions while avoiding the harmful generation of toxic species. Here, to better understand how iron levels are controlled, we carry out genome-wide mutagenesis screens in human cells. Alongside mapping known components of iron sensing, we determine the relative contributions of iron uptake, iron recycling, ferritin breakdown, and mitochondrial flux in controlling the labile iron pool. We also identify SETD2, a histone methyltransferase, as a chromatin modifying enzyme that controls intracellular iron availability through ferritin breakdown. Functionally, we show that SETD2 inhibition or cancer-associated SETD2 mutations render cells iron deficient, thereby driving resistance to ferroptosis and potentially explaining how some tumors evade antitumoral immunity.
Figures

References
-
- Rouault T. A., Hentze M. W., Caughman S. W., Harford J. B., Klausner R. D., Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science 241, 1207–1210 (1988). - PubMed
-
- Samaniego F., Chin J., Iwai K., Rouault T. A., Klausner R. D., Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. Structure, function, and post-translational regulation. J. Biol. Chem. 269, 30904–30910 (1994). - PubMed
-
- Guo B., Yu Y., Leibold E. A., Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J. Biol. Chem. 269, 24252–24260 (1994). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

