Directed-complement killing of Pseudomonas aeruginosa protects against lethal pneumonia
- PMID: 40961506
- PMCID: PMC12466147
- DOI: 10.1016/j.ebiom.2025.105926
Directed-complement killing of Pseudomonas aeruginosa protects against lethal pneumonia
Abstract
Background: Multidrug-resistant Pseudomonas aeruginosa raises major clinical concerns due to its capacity to cause a wide-array of infections in individuals with compromised immune defences and to withstand standard-of-care therapeutic treatments. Antibody-based approaches have proven to be efficient in the treatment of diverse infections. Here we propose an innovative approach harnessing the complement at the surface of bacteria for further killing.
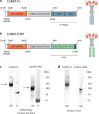
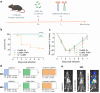
Methods: We developed two Complement-activating Multimeric immunotherapeutic compleXes (CoMiX) targeting the bacterium through a single-chain variable fragment directed against the exopolysaccharide Psl, and carrying one of two different effector functions, Factor H Related protein 1 (FHR1) or a Fc dimer. Each CoMiX was assessed in vitro for their antibacterial activity, and further evaluated in a mouse model of acute pneumonia.
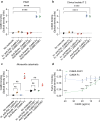
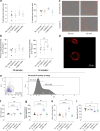
Findings: Both CoMiX-FHR1 and CoMiX-Fc effectively deposit C1q (for CoMiX-Fc), C3b, and C5b9 at the surface of multidrug-resistant clinical isolates, promoting their direct killing and/or opsonisation and subsequent phagocytosis for CoMiX-Fc (p < 0.001). Both CoMiX synergise with amikacin and protect epithelial cells against P. aeruginosa-induced cytotoxicity. Importantly, CoMiX administered intranasal to acutely infected mice significantly improve their survival (p < 0.001) by reducing local bacterial burden through the higher induction of C3b (opsonisation) and C5a (neutrophils recruitment and activation) and by decreasing lung inflammation.
Interpretation: Our proof-of-concept demonstrates the efficient, direct and indirect killing of P. aeruginosa by the complement, highlighting the therapeutic potential of CoMiX to combat multidrug-resistant bacteria.
Funding: Luxembourg National Research Fund, Ministry of Higher Education and Research of Luxembourg, COST action CA21145 EURESTOP, Institut National de la Santé et de la Recherche Médicale, and Tours University.
Keywords: Complement system; FHR1; Immunotherapy; Multidrug resistance; Pseudomonas aeruginosa.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests A patent application has been filed for CoMiX (LIH-023-PCT WO2023281120) by the inventors (B.Brandus, J.Z, X.D, C.S.D). The authors have declared that no other conflict of interest exists.
Figures





References
-
- Miller W.R., Arias C.A. ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat Rev Microbiol. 2024;22(10):598–616. - PubMed
-
- Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

