Tumor-specific but immunosuppressive CD39+CD8+ T cells exhibit double-faceted roles in clear cell renal cell carcinoma
- PMID: 40961944
- PMCID: PMC12629791
- DOI: 10.1016/j.xcrm.2025.102360
Tumor-specific but immunosuppressive CD39+CD8+ T cells exhibit double-faceted roles in clear cell renal cell carcinoma
Abstract
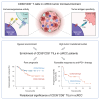
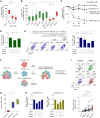
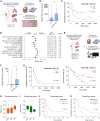
CD39+CD8+ T cells are known as tumor-antigen-specific cells among CD8+ tumor-infiltrating lymphocytes (TILs). However, CD39+CD8+ T cells also reportedly exhibit immunosuppressive activity in hypoxic tumor models. Here, we investigate CD39+CD8+ TILs in clear cell renal cell carcinoma (ccRCC), a Von Hippel-Lindau (VHL) mutation-associated hypoxic tumor. Single-cell analyses confirm that CD39+CD8+ cells are a terminally exhausted subset of tumor-specific CD8+ TILs. CD39+CD8+ T cell development is directly induced by cAMP and T cell receptor (TCR) signaling. Analysis of a renal cell carcinoma (RCC) cohort reveals that the proportion of CD39+CD8+ TILs is associated with a high tumor mutational burden and hypoxic features. Ex vivo functional assays reveal that CD39+CD8+ TILs exert immunosuppressive activity via ectonucleotidase activity- and adenosine-dependent mechanisms. CD39+CD8+ TIL enrichment predicts poor prognosis in patients with ccRCC yet also predicts favorable treatment responses to anti-programmed cell death protein 1 (PD-1) therapy. This paradoxical prognostic significance in ccRCC is explained by the dual properties of CD39+CD8+ TILs: tumor antigen specificity and immunosuppressive activity.
Keywords: CD39(+)CD8(+) T cells; adenosine pathway; anti-PD-1 therapy; clear cell renal cell carcinoma; hypoxia; immunosuppressive activity; paradoxical prognosis; tumor antigen specificity; tumor microenvironment.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

