The impact of pH on proteolytic activity in wound fluid: Implications for acid therapy
- PMID: 40962052
- PMCID: PMC12550788
- DOI: 10.1016/j.jbc.2025.110723
The impact of pH on proteolytic activity in wound fluid: Implications for acid therapy
Abstract
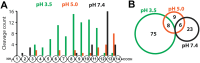
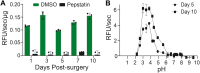
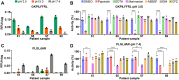
Wound healing necessitates a balance between synthesis and breakdown of extracellular matrix components, which is tightly regulated by proteases and their inhibitors. While studies have demonstrated that citric and acetic acid treatments enhance healing in recalcitrant wounds, the underlying proteolytic mechanisms remain elusive. In this study, we systematically evaluated changes in the proteolytic activity of murine wound fluid upon acidification. A library of 228 synthetic peptides served as reporters of protease activity at pH 7.4, pH 5.0, and pH 3.5. The peptide digestion patterns differed at each pH, revealing that proteases active at pH 7.4 are inactivated at pH 3.5. Notably, cathepsin D emerged as the dominant active enzyme at pH 3.5, and its activity was inhibited by pepstatin. Using a fluorogenic substrate, we quantified cathepsin D activity across varying pH levels and demonstrated optimal activity between pH 3.0 and 3.8. This activity was detectable as early as 1 day postinjury and persisted over the following 10 days. Importantly, human wound fluid exhibited the same activity profile, validating the mouse model as a relevant system for studying acid-mediated wound healing processes.
Keywords: MSP-MS; acid treatment; aspartic acid protease; fluorogenic substrate; pepstatin; proteolytic activity; wound fluid.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Nussbaum S.R., Carter M.J., Fife C.E., DaVanzo J., Haught R., Nusgart M., et al. An economic evaluation of the impact, cost, and medicare policy implications of chronic nonhealing wounds. Value Health. 2018;21:27–32. - PubMed
-
- Singh N., Bhattacharyya D. Proteases in Human Diseases. Springer; Singapore: 2017. Proteases in wound healing and immunity; pp. 147–170. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

