Genetic Insights Into Pathways Supporting Optimized Biological Nitrogen Fixation in Chickpea and Their Interaction With Disease Resistance Breeding
- PMID: 40962294
- PMCID: PMC12443505
- DOI: 10.1111/ppl.70514
Genetic Insights Into Pathways Supporting Optimized Biological Nitrogen Fixation in Chickpea and Their Interaction With Disease Resistance Breeding
Abstract
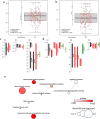
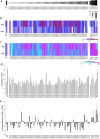
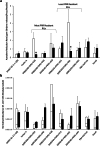
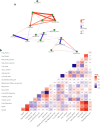
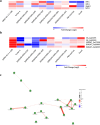
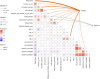
In chickpea (Cicer arietinum), a globally important grain legume, improvements in yield stability are required to address food security and agricultural land loss. One approach is to improve both nutrient acquisition through symbiosis with rhizobial bacteria and biotic stress resistance. To support the simultaneous selection of multiple beneficial traits, we sought to identify quantitative trait loci (QTL) and genes linked to improved plant-microbe symbiosis both under symbiosis-promotive growth conditions and when pathogens are present. Our aims were to use the chickpea-Mesorhizobium rhizobial model to identify QTL associated with biological nitrogen fixation (BNF) and nutrient acquisition and understand factors promotive of sustained BNF under biotic stress through the impact of Phytophthora root rot (PRR) on BNF across chickpea genotypes on host gene expression. Using two chickpea × C. echinospermum recombinant inbred line (RIL) populations, we identified QTL associated with BNF and several associated with macro- and micro-nutrient status of chickpea. From within a set of the most PRR-resistant RIL (n = 70), we successfully identified RIL with both high PRR resistance and N sourced from BNF. In conditions of the tripartite (host:rhizobia:pathogen) interaction, while there was no consistent pathogen impact on the abundance of Mesorhizobium in nodules, PRR-resistant genotypes maintained a higher activity of their N-assimilation genes, while susceptible genotypes repressed these genes. This improved understanding of the genetic support of BNF in chickpea will allow selection for material that maintains higher BNF and is more disease resistant, which together may improve yield stability in chickpea.
Keywords: disease resistance; nitrogen fixation; quantitative trait loci identification.
© 2025 The Author(s). Physiologia Plantarum published by John Wiley & Sons Ltd on behalf of Scandinavian Plant Physiology Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Abbo, S. , Redden R. J., and Yadav S. S.. 2007. “Utlization of Wild Relatives.” In Chickpea Breeding & Management, edited by Yadav S. S., Redden R. J., Chen W., and Sharma B., 338–354. CABI.
-
- Abi‐Ghanem, R. , Carpenter‐Boggs L., Smith J., and Vandemark G.. 2012. “Nitrogen Fixation by US and Middle Eastern Chickpeas With Commercial and Wild Middle Eastern Inocula.” International Scholarly Research Notices 5: 1–5.
-
- Alexa, A. , and Rahnenfuhrer J.. 2021. “topGO: Enrichment Analysis for Gene Ontology.” R Package Version 2.44.0.
-
- Amalraj, A. , Baumann U., Hayes J. E., and Sutton T.. 2024. “Using RNA Sequencing to Unravel Molecular Changes Underlying the Defense Response in Chickpea Induced by Phytophthora medicaginis .” Physiologia Plantarum 176: e14412. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

