Infiltrating macrophages replace Kupffer cells and play diverse roles in severe alcohol-associated hepatitis
- PMID: 40962953
- PMCID: PMC12480828
- DOI: 10.1038/s41423-025-01343-1
Infiltrating macrophages replace Kupffer cells and play diverse roles in severe alcohol-associated hepatitis
Abstract
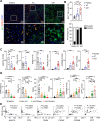
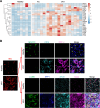
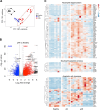
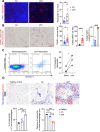
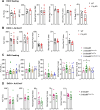
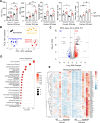
Patients with alcohol-associated cirrhosis (AC) may develop severe alcohol-associated hepatitis (sAH), a disease with high short-term mortality. Our previous studies demonstrated that sAH, but not AC livers, are infiltrated with a high number of self-sustaining IL-8+ neutrophils that likely drive the transition from AC to sAH. Monocyte-derived macrophages (MoMFs) also infiltrate the liver in sAH, but their roles remain largely obscure. In the present study, we characterized liver macrophages in human liver explants from sAH and AC patients. Our data revealed a marked reduction in Kupffer cells, whereas MoMFs were increased in sAH and AC. Single-cell RNA-Seq analyses revealed several populations in both AC and sAH, including C1Q+, S100A8+, APOE+, TNF+ and VSIG4+ macrophages, with sAH containing unique C1Q+ macrophages potentially playing a role in removing apoptotic neutrophils in sAH. C1Q+ macrophages also express many genes involved in phagocytosis and proinflammatory and anti-inflammatory functions, suggesting that C1Q+ macrophages have diverse functions in sAH. The roles of C1Q, S100A8, and APOE were further examined in experimental models of alcohol-induced liver injury. Our data revealed that C1q KO mice and macrophage-specific S100a8 KO mice presented similar alcohol-induced liver injury and hepatic neutrophil infiltration, while Apoe KO mice developed much more severe liver injury than did WT mice following chronic-plus-binge ethanol challenge. Taken together, sAH and AC are infiltrated with multiple populations of macrophages that perform diverse functions to drive chronic disease progression. Unique C1Q+ macrophages in sAH play a compensatory role in removing dead cells but may also promote inflammation in sAH.
Keywords: APOE; C1Q; Macrophages; Neutrophil apoptosis; S100A8.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests. Dr. Bin Gao is Associate Editor-in-Chief of Cellular & Molecular Immunology, but he has not been involved in the peer review or the decision-making of the article.
Figures


References
-
- Vergis N, Patel V, Bogdanowicz K, Czyzewska-Khan J, Keshinro R, Fiorentino F, et al. IL-1 Signal inhibition in alcohol-related hepatitis: a randomized, double-blind, placebo-controlled trial of Canakinumab. Clin Gastroenterol Hepatol. 2024;23:797–807. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

