CRISPR activation for SCN2A-related neurodevelopmental disorders
- PMID: 40963013
- PMCID: PMC12581945
- DOI: 10.1038/s41586-025-09522-w
CRISPR activation for SCN2A-related neurodevelopmental disorders
Erratum in
-
Publisher Correction: CRISPR activation for SCN2A-related neurodevelopmental disorders.Nature. 2026 Jan;649(8095):E2. doi: 10.1038/s41586-025-09995-9. Nature. 2026. PMID: 41367045 No abstract available.
Abstract
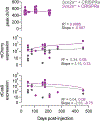
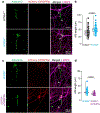
Most neurodevelopmental disorders with single gene diagnoses act via haploinsufficiency, in which only one of the two gene copies is functional1. SCN2A haploinsufficiency is one of the most frequent causes of neurodevelopmental disorder, often presenting with autism spectrum disorder, intellectual disability and, in a subset of children, refractory epilepsy2. Here, using SCN2A haploinsufficiency as a proof-of-concept, we show that upregulation of the existing functional gene copy through CRISPR activation (CRISPRa) can rescue neurological-associated phenotypes in Scn2a haploinsufficient mice. We first show that restoring Scn2a expression in adolescent heterozygous Scn2a conditional knock-in mice rescues electrophysiological deficits associated with Scn2a haploinsufficiency (Scn2a+/-). Next, using an adeno-associated virus CRISPRa-based treatment in adolescent mice, we show that we can correct intrinsic and synaptic deficits in neocortical pyramidal cells, a major cell type that contributes to neurodevelopmental disorders and seizure aetiology in SCN2A haploinsufficiency. Furthermore, we find that systemic delivery of CRISPRa protects Scn2a+/- mice against chemoconvulsant-induced seizures. Finally, we also show that adeno-associated virus CRISPRa treatment rescues excitability in SCN2A haploinsufficient human stem-cell-derived neurons. Our results showcase the potential of this therapeutic approach to rescue SCN2A haploinsufficiency and demonstrates that rescue even at adolescent stages can ameliorate neurodevelopmental phenotypes.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: N.M. is the cofounder and former board member and CSO of Regel Therapeutics, N.A. is the cofounder of Regel Therapeutics and both N.A. and K.J.B. are on the scientific advisory board of Regel Therapeutics. P.W.E.S. is a Program Director at Regel Therapeutics. N.M. and N.A. are the inventors on patent ‘Gene therapy for haploinsufficiency’ WO2018148256A9. N.A., K.J.B. and S.J.S. receive funding from BioMarin Pharmaceutical Incorporated. The other authors declare no competing interests.
Figures



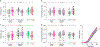







References
MeSH terms
Substances
Grants and funding
- U54 NS108874/NS/NINDS NIH HHS/United States
- P30 DK063720/DK/NIDDK NIH HHS/United States
- R00 NS078118/NS/NINDS NIH HHS/United States
- R01 MH136475/MH/NIMH NIH HHS/United States
- S10 OD021822/OD/NIH HHS/United States
- T32 GM007449/GM/NIGMS NIH HHS/United States
- F32 MH125536/MH/NIMH NIH HHS/United States
- R01 MH125978/MH/NIMH NIH HHS/United States
- R01 NS121287/NS/NINDS NIH HHS/United States
- R01 MH115045/MH/NIMH NIH HHS/United States
- K99 MH135209/MH/NIMH NIH HHS/United States
- R01 MH118298/MH/NIMH NIH HHS/United States
- K99 NS078118/NS/NINDS NIH HHS/United States
- R01 MH126960/MH/NIMH NIH HHS/United States

