Peroxisomal metabolism of branched fatty acids regulates energy homeostasis
- PMID: 40963015
- PMCID: PMC12571886
- DOI: 10.1038/s41586-025-09517-7
Peroxisomal metabolism of branched fatty acids regulates energy homeostasis
Abstract
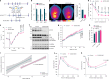
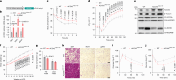
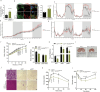
Brown and beige adipocytes express uncoupling protein 1 (UCP1), a mitochondrial protein that dissociates respiration from ATP synthesis and promotes heat production and energy expenditure. However, UCP1-/- mice are not obese1-5, consistent with the existence of alternative mechanisms of thermogenesis6-8. Here we describe a UCP1-independent mechanism of thermogenesis involving ATP-consuming metabolism of monomethyl branched-chain fatty acids (mmBCFA) in peroxisomes. These fatty acids are synthesized by fatty acid synthase using precursors derived from catabolism of branched-chain amino acids9 and our results indicate that β-oxidation of mmBCFAs is mediated by the peroxisomal protein acyl-CoA oxidase 2 (ACOX2). Notably, cold exposure upregulated proteins involved in both biosynthesis and β-oxidation of mmBCFA in thermogenic fat. Acute thermogenic stimuli promoted translocation of fatty acid synthase to peroxisomes. Brown-adipose-tissue-specific fatty acid synthase knockout decreased cold tolerance. Adipose-specific ACOX2 knockout also impaired cold tolerance and promoted diet-induced obesity and insulin resistance. Conversely, ACOX2 overexpression in adipose tissue enhanced thermogenesis independently of UCP1 and improved metabolic homeostasis. Using a peroxisome-localized temperature sensor named Pexo-TEMP, we found that ACOX2-mediated fatty acid β-oxidation raised intracellular temperature in brown adipocytes. These results identify a previously unrecognized role for peroxisomes in adipose tissue thermogenesis characterized by an mmBCFA synthesis and catabolism cycle.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: I.J.L. and X.L. are named on a provisional patent application (serial no. 63/872,889) filed by Washington University related to targeting ACOX2 activation as a treatment for obesity and related metabolic diseases. The other authors declare no competing interests.
Figures












References
-
- Dieckmann, S. et al. Susceptibility to diet-induced obesity at thermoneutral conditions is independent of UCP1. Am. J. Physiol. Endocrinol. Metabol.322, E85–E100 (2022). - PubMed
-
- Enerbäck, S. et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature387, 90–94 (1997). - PubMed
-
- Feldmann, H. M., Golozoubova, V., Cannon, B. & Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab.9, 203–209 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

