Covariation MS uncovers a protein that controls cysteine catabolism
- PMID: 40963025
- PMCID: PMC12589099
- DOI: 10.1038/s41586-025-09535-5
Covariation MS uncovers a protein that controls cysteine catabolism
Abstract
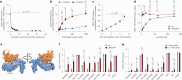
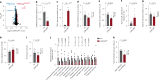
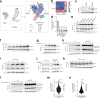
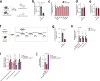
The regulation of metabolic processes by proteins is fundamental to biology and yet is incompletely understood. Here we develop a mass spectrometry (MS)-based approach that leverages genetic diversity to nominate functional relationships between 285 metabolites and 11,868 proteins in living tissues. This method recapitulates protein-metabolite functional relationships mediated by direct physical interactions and local metabolic pathway regulation while nominating 3,542 previously undescribed relationships. With this foundation, we identify a mechanism of regulation over liver cysteine utilization and cholesterol handling, regulated by the poorly characterized protein LRRC58. We show that LRRC58 is the substrate adaptor of an E3 ubiquitin ligase that mediates proteasomal degradation of CDO1, the rate-limiting enzyme of the catabolic shunt of cysteine to taurine1. Cysteine abundance regulates LRRC58-mediated CDO1 degradation, and depletion of LRRC58 is sufficient to stabilize CDO1 to drive consumption of cysteine to produce taurine. Taurine has a central role in cholesterol handling, promoting its excretion from the liver2, and we show that depletion of LRRC58 in hepatocytes increases cysteine flux to taurine and lowers hepatic cholesterol in mice. Uncovering the mechanism of LRRC58 control over cysteine catabolism exemplifies the utility of covariation MS to identify modes of protein regulation of metabolic processes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.T.C. is co-founder and equity holder of Matchpoint Therapeutics and Aevum Therapeutics. E.S.F. is a founder, scientific advisory board member and equity holder of Civetta Therapeutics, Proximity Therapeutics, Neomorph Inc. (serving on the board of directors), Stelexis Biosciences Inc., Anvia Therapeutics Inc. (serving on the board of directors) and CPD4 Inc. (serving on the board of directors). E.S.F. is also an equity holder and scientific advisory board member for Avilar Therapeutics, Photys Therapeutics and Ajax Therapeutics and an equity holder in Lighthorse Therapeutics. E.S.F. is a consultant to Novartis, EcoR1 capital, Odyssey and Deerfield. The Fischer laboratory receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer and Astellas. K.A.D. receives or has received consulting fees from Neomorph Inc. and Kronos Bio. E.T.C., H.X. and M.O. have filed patents on LRRC58. The other authors declare no competing interests.
Figures











References
-
- Stipanuk, M. H., Dominy, J. E. Jr, Lee, J. I. & Coloso, R. M. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J. Nutr.136, 1652S–1659S (2006). - PubMed
-
- Hofmann, A. F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med.159, 2647–2658 (1999). - PubMed
-
- Chubukov, V., Gerosa, L., Kochanowski, K. & Sauer, U. Coordination of microbial metabolism. Nat. Rev. Microbiol.12, 327–340 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

