This is a preprint.
De novo design of semisynthetic protein nanopores
- PMID: 40964308
- PMCID: PMC12439879
- DOI: 10.1101/2025.09.08.674823
De novo design of semisynthetic protein nanopores
Abstract
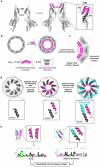
Protein nanopores are essential components of single-molecule oligonucleotide sequencing and sensing devices. Here, we demonstrate that installing additional de novo subunits enables large-scale architectural changes of nanopore complexes. We design de novo proteins that integrate seamlessly with the CsgG pore to form 18-subunit, 315-kilodalton complexes with precisely sculpted pore architectures and tailored ion conduction, opening new possibilities for engineering enhanced nanopores with customized structural and functional properties.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
