This is a preprint.
Identification of a broad and potent V3 glycan site bNAb targeting an N332gp120 glycan-independent epitope
- PMID: 40964353
- PMCID: PMC12439949
- DOI: 10.1101/2025.09.05.674437
Identification of a broad and potent V3 glycan site bNAb targeting an N332gp120 glycan-independent epitope
Update in
-
Identification of a potent V3 glycan site broadly neutralizing antibody targeting an N332gp120 glycan-independent epitope.Nat Immunol. 2026 Feb 3. doi: 10.1038/s41590-025-02385-3. Online ahead of print. Nat Immunol. 2026. PMID: 41634487
Abstract
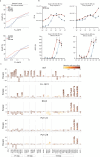
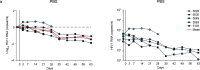
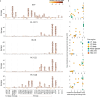
Broadly neutralizing antibodies (bNAbs) against HIV-1 can suppress viremia in vivo and inform vaccine development. Here, we characterized 007, a V3 glycan site bNAb exhibiting high levels of antiviral activity against multiclade pseudovirus panels1-3 (GeoMean IC50 = 0.012 μg/mL, breadth = 69%, 217 virus strains) by targeting a N332gp120 glycan-independent V3 epitope, a site of Env vulnerability to which only weakly neutralizing antibodies had previously been identified. Functional analyses demonstrated distinct binding and neutralization profiles compared to classical V3 glycan site bNAbs. A 007 Fab-Env cryo-EM structure revealed contacts with the V3 324GD/NIR327 motif and interactions with N156gp120 and N301gp120 glycans. In contrast to classical V3 bNAbs, 007 binding to Env does not depend on the N332gp120 glycan, rendering it resistant to common escape mutations. Structures of 007 IgG-Env trimer complexes showed two Env trimers crosslinked by three bivalent IgGs, and bivalent 007 IgG was up to ~300-fold more potent than monovalent 007 IgG heterodimer, suggesting a role for avidity in potent neutralization. Finally, in HIV-1ADA-infected humanized mice, 007 caused transient decline of viremia and overcame classical V3 escape mutations, highlighting 007's potential for HIV-1 prevention, therapy, functional cure, and vaccine design.
Conflict of interest statement
Competing interest declaration A patent application incorporating elements of this study has been submitted by the University of Cologne, with L.G. and F.K. listed as inventors. Additionally, H.G., P.S., and F.K. are named inventors on other patent applications related to HIV-1 neutralizing antibodies. L.G., H.G., P.F.S., and F.K. have received financial compensation from the University of Cologne for licensed patents. C.H.D, S.D, F.G., D.C. are employees and stockholders of Vir Biotechnology, Inc. J.D.B. and C.R. are inventors on patents related to viral deep mutational scanning, licensed by Fred Hutch. J.D.B. also serves as a consultant for Apriori Bio. MBR and JN are co-founders and co-owners of Reliant Glycosciences, LLC, Birmingham, AL, USA.
Figures
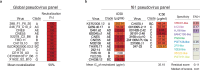












References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
