This is a preprint.
Structure-based discovery of inhibitors of Mac1 domain of nonstructural protein-3 of SARS-CoV-2 by machine learning-augmented screening of chemical space
- PMID: 40964377
- PMCID: PMC12440007
- DOI: 10.1101/2025.09.05.674529
Structure-based discovery of inhibitors of Mac1 domain of nonstructural protein-3 of SARS-CoV-2 by machine learning-augmented screening of chemical space
Abstract
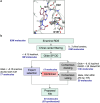
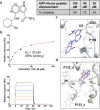
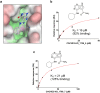
Significant efforts have been recently dedicated to the discovery of small molecule inhibitors against the Macrodomain 1 (Mac1) of nonstructural protein 3 (NSP3) as potential antivirals for SARS-CoV-2. Thus, Mac1 has also been selected as the target for the Critical Assessment of Hit-finding Experiments (CACHE) challenge #3. As contestants in that challenge, we developed a computational strategy that ranked on the top among all 23 participants in the competition and resulted in the discovery of a novel chemical series of non-charged Mac1 inhibitors. Those have been identified through the combination of machine learning-accelerated virtual screening of Enamine REAL Diversity Subset of approximately 25 million compounds and consequent hit expansion into the entire Enamine REAL Space library. In particular, the initially identified hit compound CACHE3-HI_1706_56 (KD = 20 μM) was explored by probing 17 close analogues from a library of 44 billion molecules from the Enamine REAL. All those analogues effectively displaced the Mac1-binding ADP-ribose peptide, and 12 were confirmed to engage with Mac1 by the Surface Plasmon Resonance experiments, revealing a new chemical series of compounds for hit-to-lead optimization. The structure of the CACHE3-HI_1706_56-Mac1 complex was further determined at high resolution with crystallography, confirming initial computational predictions. Our results illustrate the effectiveness of ML-accelerated docking to rapidly identify novel chemical series and provide a strong foundation for the development of SARS-CoV-2 NSP3 Mac1 inhibitors.
Keywords: CACHE challenge; Mac1; SARS-CoV-2; Virtual screening; machine learning.
Conflict of interest statement
Conflicts of Interest FG is a co-founder and advisor of In Virtuo Laboratories. JSF is a consultant to and a shareholder of Vilya Therapeutics and Relay Therapeutics. These companies had no role in the design or conduct of the study; in the collection, analysis, or interpretation of the data; or in the preparation, review, or approval of the manuscript.
Figures

References
-
- Alhammad Y. M. O.; Kashipathy M. M.; Roy A.; Gagne J. P.; McDonald P.; Gao P.; Nonfoux L.; Battaile K. P.; Johnson D. K.; Holmstrom E. D.; Poirier G. G.; Lovell S.; Fehr A. R., The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase. J Virol 2021, 95 (3).
-
- Grunewald M. E.; Chen Y.; Kuny C.; Maejima T.; Lease R.; Ferraris D.; Aikawa M.; Sullivan C. S.; Perlman S.; Fehr A. R., The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog 2019, 15 (5), e1007756. - PMC - PubMed
-
- Correy Galen J., K. D. W., Phillips Gwyndalyn, Pant Swati, Russi Silvia,; Cohen Aina E., M. G., Holton James M., Gahbauer Stefan, Thompson Michael C.,; Ashworth Alan, C. L., Kovalevsky Andrey, Meilleur Flora, Fraser James S., The mechanisms of catalysis and ligand binding for the SARS-CoV-2 NSP3 macrodomain from neutron and x-ray diffraction at room temperature. Sci. Adv. 2022, 8, (eabo5083), 1–19.
-
- Russo L. C.; Tomasin R.; Matos I. A.; Manucci A. C.; Sowa S. T.; Dale K.; Caldecott K. W.; Lehtio L.; Schechtman D.; Meotti F. C.; Bruni-Cardoso A.; Hoch N. C., The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J Biol Chem 2021, 297 (3), 101041. - PMC - PubMed
-
- Fehr A. R.; Channappanavar R.; Jankevicius G.; Fett C.; Zhao J.; Athmer J.; Meyerholz D. K.; Ahel I.; Perlman S., The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2016, 7 (6).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
