PTBP1 variants displaying altered nucleocytoplasmic distribution are responsible for a neurodevelopmental disorder with skeletal dysplasia
- PMID: 40965981
- PMCID: PMC12618068
- DOI: 10.1172/JCI182100
PTBP1 variants displaying altered nucleocytoplasmic distribution are responsible for a neurodevelopmental disorder with skeletal dysplasia
Abstract
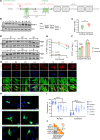
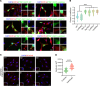
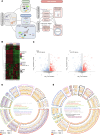
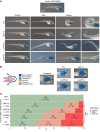
Polypyrimidine tract-binding protein 1 (PTBP1) is a heterogeneous nuclear ribonucleoprotein primarily known for its alternative splicing activity. It shuttles between the nucleus and cytoplasm via partially overlapping N-terminal nuclear localization (NLS) and export (NES) signals. Despite its fundamental role in cell growth and differentiation, its involvement in human disease remains poorly understood. We identified 27 individuals from 25 families harboring de novo or inherited pathogenic variants - predominantly start-loss (89%) and, to a lesser extent, missense (11%) - affecting NES/NLS motifs. Affected individuals presented with a syndromic neurodevelopmental disorder and variable skeletal dysplasia with disproportionate short stature with short limbs. Intellectual functioning ranged from normal to moderately delayed. Start-loss variants led to translation initiation from an alternative downstream in-frame methionine, resulting in loss of the NES and the first half of the bipartite NLS, and increased cytoplasmic stability. Start-loss and missense variants shared a DNA methylation episignature in peripheral blood and altered nucleocytoplasmic distribution in vitro and in vivo with preferential accumulation in processing bodies, causing aberrant gene expression but normal RNA splicing. Transcriptomic analysis of patient-derived fibroblasts revealed dysregulated pathways involved in osteochondrogenesis and neurodevelopment. Overall, our findings highlight a cytoplasmic role for PTBP1 in RNA stability and disease pathogenesis.
Keywords: Bone development; Development; Genetic diseases; Genetics; RNA processing.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

