Disrupting integrator complex subunit INTS6 causes neurodevelopmental disorders and impairs neurogenesis and synapse development
- PMID: 40966122
- PMCID: PMC12618080
- DOI: 10.1172/JCI191729
Disrupting integrator complex subunit INTS6 causes neurodevelopmental disorders and impairs neurogenesis and synapse development
Abstract
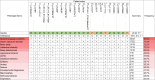
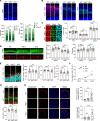
The Integrator complex plays essential roles in RNA polymerase II (RNAPII) transcription termination and RNA processing. Here, we identify INTS6, a subunit of the Integrator complex, as a novel gene associated with neurodevelopmental disorders (NDDs). Through analysis of large NDD cohorts and international collaborations, we identified 23 families harboring monoallelic likely gene-disruptive or de novo missense variants in INTS6. Phenotypic characterization revealed shared features, including language and motor delays, autism, intellectual disability, and sleep disturbances. Using a nervous-system conditional KO (cKO) mouse model, we show that Ints6 deficiency disrupts early neurogenesis, cortical lamination, and synaptic development. Ints6 cKO mice had a thickened ventricular zone/subventricular zone, thinning of the cortical plate, reduced neuronal differentiation, and increased apoptosis in cortical layer 6. Behavioral assessments of heterozygous mice revealed deficits in social novelty preference, spatial memory, and hyperactivity, mirroring phenotypes observed in individuals with INTS6 variants. Molecular analyses further revealed that INTS6 deficiency alters RNAPII dynamics, disrupts transcriptional regulation, and impairs synaptic gene expression. Treatment with a CDK9 inhibitor (CDK9i) reduced RNAPII phosphorylation, thereby limiting its binding to target genes. Notably, CDK9i reversed neurosphere overproliferation and rescued the abnormal dendritic spine phenotype caused by Ints6 deficiency. This work advances understanding of INTS-related NDD pathogenesis and highlights potential therapeutic targets for intervention.
Keywords: Genetic variation; Genetics; Neuroscience.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

