State-by-state influenza outbreaks and oversee: A Markov chain study of California and North Carolina, USA
- PMID: 40966267
- PMCID: PMC12445519
- DOI: 10.1371/journal.pgph.0005135
State-by-state influenza outbreaks and oversee: A Markov chain study of California and North Carolina, USA
Abstract
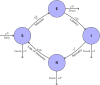
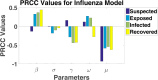
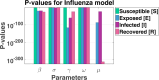
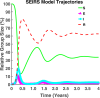
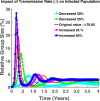
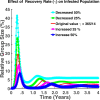
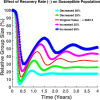
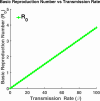
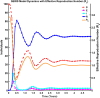
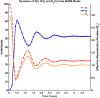
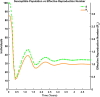
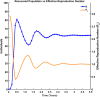
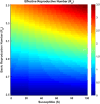
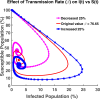
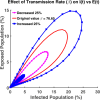
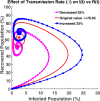
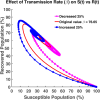
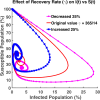
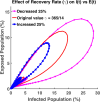
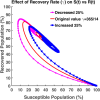
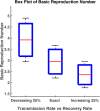
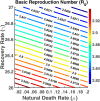
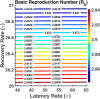
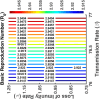
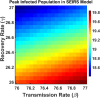
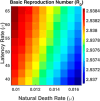
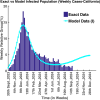
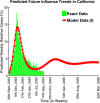
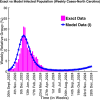
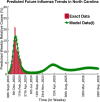
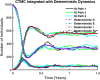
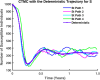
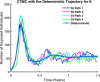
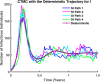
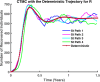
Influenza, a significant public health concern, spreads rapidly and causes seasonal epidemics and pandemics. Mathematical models are essential tools for devising effective strategies to combat this pandemic. Various models have been utilized to study influenza's transmission dynamics and control measures. This paper presents the SEIRS (Susceptible-Exposed-Infectious-Recovered-Susceptible) model to analyze the disease's transmission dynamics. The model analyzes real data from California and North Carolina to assess trends, identify key factors, and project the nationwide spread of the disease. Subsequently, we calculate the basic reproduction number ([Formula: see text]) using the next-generation matrix method. Sensitivity analysis using Latin Hypercube Sampling (LHS) has been conducted to identify the model's most influential parameters. We graphically demonstrate how different parameters affect the exposed and infected populations, as well as the variation in the basic reproduction number with changes in parameters. We illustrate the interconnected behavior of the effective reproduction number alongside the different compartments and the basic reproduction number. We use phase plane analysis to examine the relationship between two compartments under varying parameters. Visual tools like boxplots, contour plots, and heat maps provide insights into how different factors influence the basic reproduction number and disease transmission. We investigate the stochastic behavior of the model by transforming it into a Continuous-Time Markov Chain (CTMC) model and visualizing the results graphically. We apply the SEIRS model to real influenza data, showcasing its effectiveness in analyzing transmission dynamics, predicting outbreaks, and evaluating public health strategies for better epidemic management.
Copyright: © 2025 Akhi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures













References
-
- Yang KH, Hsu JY. A new SIR-based model for influenza epidemic. Int J Health Med Eng. 2012;6(7):701–6.
LinkOut - more resources
Full Text Sources
