ComFB, a widespread family of c-di-NMP receptor proteins
- PMID: 40966295
- PMCID: PMC12448109
- DOI: 10.1073/pnas.2513041122
ComFB, a widespread family of c-di-NMP receptor proteins
Abstract
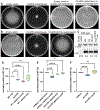
Cyclic dimeric-GMP (c-di-GMP) is a ubiquitous bacterial second messenger that regulates a variety of cellular processes, including motility, biofilm formation, secretion, cell cycle progression, and development, and also contributes to the virulence of many bacterial pathogens. While the genes encoding c-di-GMP cyclases and hydrolases are readily identifiable in microbial genomes, known c-di-GMP receptor domains are quite few, with only PilZ and MshEN broadly distributed across bacterial phyla. Recently, a new c-di-GMP receptor, named CdgR or ComFB, has been identified in cyanobacteria and shown to regulate cell size and natural competence. We demonstrated that CdgR proteins exhibit sequence and structural similarity to the Bacillus subtilis late competence development protein ComFB, a conserved protein of unknown function associated with bacterial competence. This prompted us to hypothesize that ComFB and ComFB-like proteins could also serve as c-di-GMP receptors. Here, we comprehensively investigated the ComFB protein family and demonstrated that ComFB proteins are evolutionarily widespread among bacteria and function as a novel family of c-di-GMP receptors. We showed that ComFB proteins from Gram-positive bacteria (B. subtilis, Thermoanaerobacter brockii) and Gram-negative pathogens (Vibrio cholerae, Treponema denticola) bind c-di-GMP with high affinity. Several ComFB proteins also bind cyclic di-adenosine monophosphate (c-di-AMP), suggesting that ComFB represents a widely distributed bacterial protein family with dual specificity for c-di-GMP and c-di-AMP. Our physiological studies further showed that ComFB plays vital roles in controlling motility in a c-di-GMP-dependent manner in two phylogenetically distant bacteria, B. subtilis and the gram-negative Shewanella oneidensis, attesting to the biological relevance of ComFB as a c-di-GMP binding protein.
Keywords: ComFB superfamily; bacterial motility; c-di-AMP signaling; c-di-GMP signaling.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
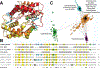

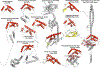

Update of
-
ComFB, a new widespread family of c-di-NMP receptor proteins.bioRxiv [Preprint]. 2024 Nov 10:2024.11.10.622515. doi: 10.1101/2024.11.10.622515. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Sep 23;122(38):e2513041122. doi: 10.1073/pnas.2513041122. PMID: 39574629 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

