The nuclear pore complex acts as a hub for pri-miRNA transcription and processing in plants
- PMID: 40966524
- PMCID: PMC12448844
- DOI: 10.1093/nar/gkaf885
The nuclear pore complex acts as a hub for pri-miRNA transcription and processing in plants
Abstract
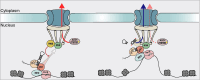
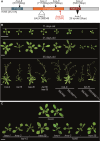
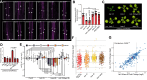
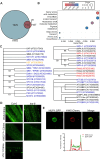
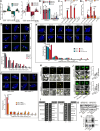
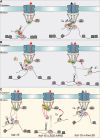
The regulation of miRNA biogenesis and movement is essential for plant development and environmental responses. HASTY (HST), a karyopherin protein, has been implicated in miRNA biogenesis and movement, though its role in non-cell-autonomous miRNA movement remains unclear. Through a genetic screen, we identified that mutations in the HAWAIIAN SKIRT (HWS) gene suppress the developmental defects of hst mutants by restoring miRNA movement. Our findings show that HWS interacts with nuclear transport factors and nuclear pore complex (NPC) components, including NUP1, positioning HWS as a regulator of miRNA nuclear export. Using microscopy and fluorescence in situ hybridization, we showed that pri-miRNA transcription, and likely their co-transcriptional processing, occur at the nuclear pore. Notably, we uncovered an antagonistic relationship between HST and HWS in regulating MIRNA transcription at the NPC and AGO1 loading, which could explain the observed changes in miRNA movement. HST promotes the association of MIRNA loci with the NPC, spatially positioning co-transcriptional processing by the NPC. Conversely, HWS negatively regulates this process by degrading MEDIATOR 37 subunits and detaching the processing complex from the NPC. Our data provide evidence of spatial coordination of miRNA transcription, biogenesis, and movement, highlighting a novel role for the NPC in the miRNA pathway.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
- PICT-2020-00757/Agencia Nacional de Promoción Científica y Tecnológica
- PICT-2021-I-A-00452/Agencia Nacional de Promoción Científica y Tecnológica
- CAI + D 2020/Universidad Nacional del Litoral
- PID2022-137037NB-I00/Spanish Ministry of Science and Innovation
- CNS2023-145312/Spanish Ministry of Science and Innovation
LinkOut - more resources
Full Text Sources

