A compendium of synthetic lethal gene pairs defined by extensive combinatorial pan-cancer CRISPR screening
- PMID: 40968372
- PMCID: PMC12445041
- DOI: 10.1186/s13059-025-03737-w
A compendium of synthetic lethal gene pairs defined by extensive combinatorial pan-cancer CRISPR screening
Abstract
Background: Synthetic lethal interactions are attractive therapeutic candidates as they enable selective targeting of cancer cells in which somatic alterations have disrupted one member of a synthetic lethal gene pair while leaving normal tissues untouched, thus minimising off-target toxicity. Despite this potential, the number of well-established and validated synthetic lethal gene pairs is modest.
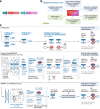
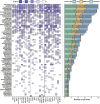
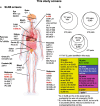
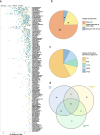
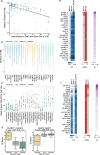
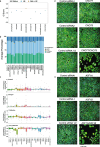
Results: We generate a dual-guide CRISPR/Cas9 Library and analyse 472 predicted synthetic lethal pairs in 27 cancer cell Lines from melanoma, pancreatic and lung cancer Lineages. We report a robust collection of 117 genetic interactions within and across cancer types and explore their candidacy as therapeutic targets. We show that SLC25A28 is an attractive target since its synthetic lethal paralog partner SLC25A37 is homozygously deleted pan-cancer. We generate knockout mice for Slc25a28 revealing that, except for cataracts in some mice, these animals are normal; suggesting inhibition of SLC25A28 is unlikely to be associated with profound toxicity.
Conclusions: We provide and validate an extensive collection of synthetic lethal interactions across cancer types.
Keywords: CRISPR; Epistasis; High-throughput screening; Synthetic lethality.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All cell lines used in this study were ethically sourced from the American Type Culture Collection except for CO92 cells which were obtained from Prof. Nick Hayward (QIMR Berghofer) with informed consent of the patient. Mouse experiments at the Wellcome Sanger Institute were performed with Home Office Approval and following approval by the Sanger Animal Welfare Committee. Competing interests: SP consults for Relation Therapeutics on studies unrelated to this work. No other authors declare competing interests.
Figures

References
-
- Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

