A Multi-Scale Mechanistic Model of Ulcerative Colitis to Investigate the Effects of Selective Suppression of IL-6 Trans-Signaling
- PMID: 40968690
- PMCID: PMC12446697
- DOI: 10.1111/cts.70366
A Multi-Scale Mechanistic Model of Ulcerative Colitis to Investigate the Effects of Selective Suppression of IL-6 Trans-Signaling
Abstract
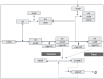
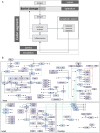
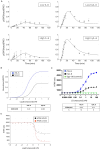
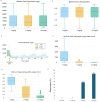
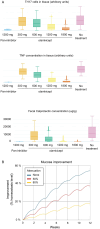
Interleukin 6 (IL-6) has previously been identified as playing a role in ulcerative colitis (UC) by activating the signal-transducing element gp130 through ligation of either the membrane-bound or soluble IL-6 receptor (termed classic and trans-signaling respectively). It has been proposed that selective inhibition of trans-IL-6 signaling could ameliorate the deleterious, pro-inflammatory effects of IL-6, while preserving the homeostatic activity of classic IL-6 signaling. We developed an in silico, mechanistic model of UC in two stages to compare the biological effects that result from inhibition of classic and trans-IL-6 signaling. In the first stage, we developed a limited-scope model of IL-6 signaling to establish the quantitative properties of classic and trans-signaling pathways on a short timescale following stimulation with IL-6. The model included both a pan-inhibitor of IL-6 classic and trans-signaling and a soluble gp130-Fc that selectively inhibited trans-signaling. In the second stage, we developed a multi-scale model of UC to study the pharmacodynamic effects of cytokine signaling inhibition and optimize treatment regimens. Across three virtual experiments, both selective and global suppression of IL-6 signaling were associated with a transition away from an inflammatory state in patients with moderate to severe inflammatory activity. In our multi-scale model, we identified a dose-response relationship between selective inhibition of trans-IL-6 signaling and tissue regeneration. Moreover, selective inhibition of trans-IL-6 signaling effectively suppressed inflammation and induced faster gut tissue healing than global IL-6 suppression. These findings suggest that global suppression of IL-6 signaling could negatively affect IL-6-induced regeneration activity, whereas this effect is less likely for selective inhibition.
Keywords: computational biology; gastrointestinal; immunology; inflammation; mechanism‐based pharmacokinetics‐pharmacodynamics; multiscale modeling.
© 2025 The Author(s). Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
O.S. was an employee of Ferring Pharmaceuticals, the sponsor of this study, at the time of this work, and is now an employee of Novo Nordisk. N.P. is an employee of PricewaterhouseCoopers A. Ravi is an employee of Ferring Pharmaceuticals, the sponsor of this study. R.C. was an employee of PricewaterhouseCoopers at the time of the study. A. Rivollier is an employee of Ferring Pharmaceuticals, the sponsor of this study. Z.W. is an employee of PricewaterhouseCoopers. S.P.V. was an employee of PricewaterhouseCoopers at the time of this work. He is now an employee of AstraZeneca and owns AstraZeneca stocks or stock options. M.B. is an employee of PricewaterhouseCoopers. S.R. is an employee of Ferring Pharmaceuticals, the sponsor of this study R.S. is an employee of Ferring Pharmaceuticals, the sponsor of this study. J.S. is an employee of PricewaterhouseCoopers. P.M.D. was a partner at PricewaterhouseCoopers at the time of this work. P.P. is an employee of Ferring Pharmaceuticals, the sponsor of this study.
Figures
References
-
- Wagner F. D., Schreiber S., Bagger Y., et al., “Safety, Tolerability, and Pharmacokinetics of Single‐ and Multiple‐Ascending Doses of Olamkicept: Results From Randomized, Placebo‐Controlled, First‐In‐Human Phase I Trials,” Clinical and Translational Science 17, no. 5 (2024): e13832, 10.1111/cts.13832. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

