Nipah virus matrix protein uses cortical actin to stabilize the virus assembly sites and promote budding
- PMID: 40971433
- PMCID: PMC12448087
- DOI: 10.1126/sciadv.adw4609
Nipah virus matrix protein uses cortical actin to stabilize the virus assembly sites and promote budding
Abstract
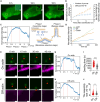
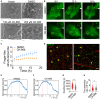
Several enveloped viruses, including paramyxoviruses, assemble and bud from the host plasma membrane (PM). Nipah virus (NiV), a deadly zoonotic paramyxovirus, uses its matrix protein (M) to drive virus assembly and budding through dimerization and PM interaction. We show that NiV-M-mediated virus-like particle (VLP) production depends on its interaction with host F-actin via its carboxyl-terminal domain. We demonstrate that F-actin retains NiV-M assembly sites at the PM by analyzing NiV-M assembly kinetics. Disrupting actin dynamics or NiV-M-actin interaction alters M nanoscale organization and reduces membrane retention, without affecting initial recruitment. We also show that the Arp2/3 complex, an actin-branching factor, promotes VLP production. Inhibiting Arp2/3 reduces NiV-M retention at the PM and impairs protrusion formation while leaving the assembly rate unchanged. These findings suggest that the host F-actin retains NiV assembly sites on the PM and promotes virus budding via Arp2/3-driven actin branching.
Figures




References
-
- D. M. Knipe, P. M. Howley, Fields’ Virology (Lippincott Williams & Wilkins, 2007).
-
- Abedi G. R., Prill M. M., Langley G. E., Wikswo M. E., Weinberg G. A., Curns A. T., Schneider E., Estimates of parainfluenza virus-associated hospitalizations and cost among children aged less than 5 years in the United States, 1998-2010. J. Pediatric Infect. Dis. Soc. 5, 7–13 (2016). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

