Drug and Clinical Candidate Drug Data in ChEMBL
- PMID: 40971497
- PMCID: PMC12516679
- DOI: 10.1021/acs.jmedchem.5c00920
Drug and Clinical Candidate Drug Data in ChEMBL
Abstract
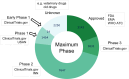
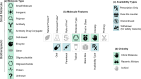
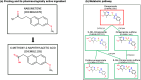
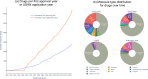
ChEMBL is a large-scale, open-access, FAIR database of bioactive molecules with drug-like properties. ChEMBL 35 contains 17,500 approved drugs, and drugs that are progressing through the clinical development pipeline. Drug curation has formed an integral part of the core offering of the ChEMBL database since its inception. The paper is a reference guide to present the principles of why the ChEMBL drug data has been curated in a particular manner so that data users can better understand the nature of the data. The drug data include information on: names, synonyms and trade names, chemical structure or biological sequence, data sources, indications, mechanisms, warnings and drug properties such as maximum phase of development, type of molecule, prodrug status and first approval. The integrated nature of the drug data within the context of a bioactivity resource enables the wide use of the data set in drug discovery, AI and machine learning.
Figures

References
-
- Global Biodata Coalition . Global Core Biodata Resource https://globalbiodata.org/what-we-do/global-core-biodata-resources (accessed 31st March 2025).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

