Hydroxyl and Trifluoromethyl Radical Carbohydrate Footprinting for Probing Protein Binding Components of Oligosaccharides
- PMID: 40973028
- PMCID: PMC12560149
- DOI: 10.1021/acs.analchem.5c02719
Hydroxyl and Trifluoromethyl Radical Carbohydrate Footprinting for Probing Protein Binding Components of Oligosaccharides
Abstract
Carbohydrates are found in various forms in living organisms, both as free-standing glycans as well as glycoconjugates including glycoproteins, glycolipids, and glycosaminoglycans. These structures play crucial roles in many biological processes, often mediated or influenced by interactions of carbohydrates with other biomolecules. However, studying these interactions is particularly challenging due to the structural complexity of carbohydrates, their dynamic conformational behavior, and the low binding affinities often involved. To address these challenges, we are developing a novel method that leverages mass spectrometry-based radical carbohydrate footprinting (RCF). We monitored changes in the solvent accessibility of specific regions within oligosaccharides by measuring variations in the apparent rate of hydroxyl radical and trifluoromethyl radical-mediated oxidation. In our studies, a collection of trisaccharide isomers and N,N',N″-triacetylchitotriose (NAG3) shows no significant change in modification in nonbinding protein solutions. However, in the presence of two proteins that bind NAG3 specifically, NAG3 oxidation is reduced. We find that the free reducing end is the primary site of hydroxyl radical oxidation under covalent labeling conditions, allowing it to distinguish interactions at the glycan reducing end. Trifluoromethyl radicals, conversely, label broadly across the trisaccharide by substitution into a C-H bond. Overall, this approach offers a powerful novel approach for identifying glycan-protein interactions and mapping the binding interface of glycans.
Figures
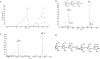
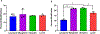
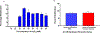
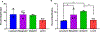


Update of
-
Hydroxyl and Trifluoromethyl Radical Carbohydrate Footprinting for Probing Protein Binding Components of Oligosaccharides.bioRxiv [Preprint]. 2025 May 10:2025.05.06.652515. doi: 10.1101/2025.05.06.652515. bioRxiv. 2025. Update in: Anal Chem. 2025 Sep 30;97(38):20809-20816. doi: 10.1021/acs.analchem.5c02719. PMID: 40654815 Free PMC article. Updated. Preprint.
References
-
- Berg JM; Tymoczko JL; Stryer L Biochemistry, 8th ed.; W.H. Freeman: New York, 2015.
-
- Pratt CW; Cornely K Essential Biochemistry; Wiley: Hoboken, NJ, 2014.
-
- Varki A; Cummings RD; Esko JD; Stanley P; Hart GW; Aebi M; Seeberger PH, et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2017.
-
- Palmer RA; Niwa H X-ray Crystallographic Studies of Protein–Ligand Interactions. Biochem. Soc. Trans 2003, 31 (5), 973–979. - PubMed
-
- Kogelberg H; Solis D; Jimenez-Barbero J New Structural Insights into Carbohydrate–Protein Interactions from NMR Spectroscopy. Curr. Opin. Struct. Biol 2003, 13 (5), 646–653. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

