AQuA Tools: clear and reliable BEDPE operations for 3D genomics
- PMID: 40973037
- PMCID: PMC12453685
- DOI: 10.1093/bioinformatics/btaf510
AQuA Tools: clear and reliable BEDPE operations for 3D genomics
Abstract
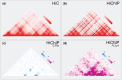
Motivation: The genome interacts with itself within the volume of the cell nucleus to process information. These interactions mediate signal integration, gene regulation, and cell identity. The identification of new therapeutic targets from non-coding disease-associated variants relies critically on correctly assigning variants to genes through 3D interactions. Experimental techniques in 3D genomics, such as HiC and HiChIP, allow the mapping of interactions through sequencing. Bioinformatics for 3D genomics contends primarily with contact matrices that contain interaction frequencies for all possible element pairs, and BEDPE files that store element pairs that interact. Whereas the tools available for processing linear genomic data are mature, operating on contact matrices and BEDPE files remains cumbersome, opaque, and error-prone, as researchers have had to shoehorn tools originally designed for linear data. A genome arithmetic designed from the ground up for 3D genomics does not yet exist.
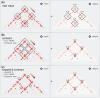
Results: We present AQuA Tools, a suite of shell- and R-based command-line tools that provide a set of core operations on contact matrices and BEDPE files motivated by key questions in population genetics, cancer research, and precision medicine. We have designed our core operations to be clear, reliable, intuitive and versatile. Core operations can be chained together along with standard UNIX commands. Our goal is to make AQuA Tools easy for the novice to learn and the go-to choice for power users. We hope our tools will motivate more researchers to use 3D genomic data in their projects.
Availability and implementation: We provide and maintain AQuA Tools at https://github.com/axiotl/aqua-tools.
© The Author(s) 2025. Published by Oxford University Press.
Figures





References
-
- Ewels PA, Peltzer A, Fillinger S et al. The Nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38:276–8. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

