Fructose and glucose from sugary drinks enhance colorectal cancer metastasis via SORD
- PMID: 40973819
- PMCID: PMC12552132
- DOI: 10.1038/s42255-025-01368-w
Fructose and glucose from sugary drinks enhance colorectal cancer metastasis via SORD
Abstract
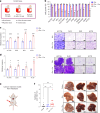
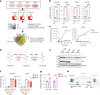
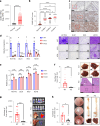
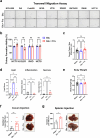
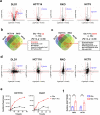
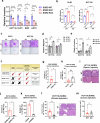
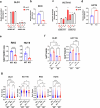
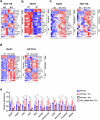
The consumption of sugar-sweetened beverages (SSBs), which contain high levels of fructose and glucose, has been causally and mechanistically linked to an increased risk of colorectal cancer (CRC). However, the effects of SSB consumption on advanced stages of disease progression, including metastasis, remain poorly understood. Here we show that exposure of CRC cells to a glucose and fructose formulation-reflecting the composition of both high-fructose corn syrup and sucrose found in SSBs-enhances cellular motility and metastatic potential compared to glucose alone. Given that CRC cells grow poorly in fructose alone, and cells in vivo are not physiologically exposed to fructose without glucose, we excluded the fructose-only condition from our studies unless needed as a control. Mechanistically, the combination of glucose and fructose elevates the NAD⁺/NADH ratio by activation of the reverse reaction of sorbitol dehydrogenase in the polyol pathway. This redox shift relieves NAD⁺ limitations and accelerates glycolytic activity, which in turn fuels activation of the mevalonate pathway, ultimately promoting CRC cell motility and metastasis. Our findings highlight the detrimental impact of SSBs on CRC progression and suggest potential dietary and therapeutic strategies to mitigate metastasis in patients with CRC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Johnson, R. J. et al. Potential role of sugar (fructose) in the epidemic of hypertension, obesity and the metabolic syndrome, diabetes, kidney disease, and cardiovascular disease. Am. J. Clin. Nutr.86, 899–906 (2007). - PubMed
-
- Bleich, S. N., Wang, Y. C., Wang, Y. & Gortmaker, S. L. Increasing consumption of sugar-sweetened beverages among US adults: 1988–1994 to 1999–2004. Am. J. Clin. Nutr.89, 372–381 (2009). - PubMed
-
- Rosinger, A., Herrick, K., Gahche, J. & Park, S. Sugar-sweetened beverage consumption among US adults, 2011–2014. NCHS Data Brief270, 1–8 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- Pew-Stewart Scholar/Pew Charitable Trusts
- R21CA258134/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- V Sholar/V Foundation for Cancer Research (V Foundation)
- P01 CA120964/CA/NCI NIH HHS/United States
- R01CA262437/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

