Genetic architecture of plasma metabolome in 254,825 individuals
- PMID: 40973927
- PMCID: PMC12449471
- DOI: 10.1038/s41467-025-62126-w
Genetic architecture of plasma metabolome in 254,825 individuals
Abstract
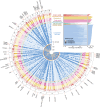
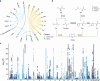
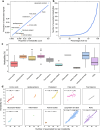
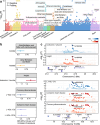
Circulating metabolites are crucial to biological processes underlying health and diseases, yet their genetic determinants remain incompletely understood. Here, we investigate the genetic architecture of nuclear magnetic resonance-based metabolomics, analyzing 249 metabolic measures and 64 biologically plausible ratios in 254,825 participants. We conduct a genome-wide association study (GWAS) identifying 24,438 independent variant-metabolite associations across 427 loci, with effect sizes highly concordant with 19 previous studies. Fine-mapping pinpoints 3610 putative causal associations, 785 of which are novel. Additionally, we utilize whole exome sequencing data and uncover 2948 gene-metabolite associations through aggregate testing, underscoring the importance of rare coding variants overlooked in GWAS. Integrating our findings with disease genetics reveals potential causal associations, such as between acetate levels and the risk of atrial fibrillation and flutter. Collectively, this study delineates the complex genetic architecture of the plasma metabolome, offering a valuable resource for future investigations into disease mechanisms and therapeutic strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

