A comprehensive landscape of AI applications in broad-spectrum drug interaction prediction: a systematic review
- PMID: 40973960
- PMCID: PMC12447627
- DOI: 10.1186/s13321-025-01093-2
A comprehensive landscape of AI applications in broad-spectrum drug interaction prediction: a systematic review
Abstract
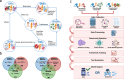
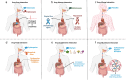
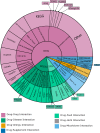
In drug development, managing interactions such as drug-drug, drug-disease, and drug-nutrient is critical for ensuring the safety and efficacy of pharmacological treatments. These interactions often overlap, forming a complex, interconnected landscape that necessitates accurate prediction to improve patient outcomes and support evidence-based care. Recent advances in artificial intelligence (AI), powered by large-scale datasets (e.g., DrugBank, TWOSIDES, SIDER), have significantly enhanced interaction prediction. Machine learning, deep learning, and graph-based models show great promise, but challenges persist, including data imbalance, noisy sources, Limited explainability, and underrepresentation of certain types of interactions. This systematic review of 147 studies (2018-2024) is the first to comprehensively map AI applications across major interaction types. We present a detailed taxonomy of models and datasets, emphasizing the growing roles of large language models and knowledge graphs in overcoming key limitations. Their integration-alongside explainable AI tools-enhances transparency, paving the way for AI-driven systems that proactively mitigate adverse interactions. By identifying the most promising approaches and critical research gaps, this review lays the groundwork for advancing more robust, interpretable, and personalized models for drug interaction prediction.
Keywords: Artificial intelligence; Deep learning; Drug–disease interactions; Drug–drug interactions; Drug–nutrient interactions; Graph-based method; Machine learning; Multiple drug interactions.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Competing interests: The authors declare no competing interests. Data and materials: All data used during this study are included in this published article (and its supplementary information files).
Figures




References
-
- Al-Anazi FHM, Talhah AFMB, Al-Anazi YAM, Alanazi NKA, Almubarak AAS, Almutairi AQK (2022) Drug interactions and their implications for patient safety. J Popul Ther Clin Pharmacol 29(04):2585–2587
-
- Jeong E et al (2024) Discovering clinical drug-drug interactions with known pharmacokinetics mechanisms using spontaneous reporting systems and electronic health records. J Biomed Inform 153:104639 - PubMed
-
- Kommu S, Carter C, Whitfield P. Adverse drug reactions. In: StatPearls. Treasure Island (FL); 2024. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

