Transcriptome-wide Mendelian randomization exploring dynamic CD4+ T cell gene expression in colorectal cancer development
- PMID: 40974097
- PMCID: PMC12571111
- DOI: 10.1093/jleuko/qiaf131
Transcriptome-wide Mendelian randomization exploring dynamic CD4+ T cell gene expression in colorectal cancer development
Abstract
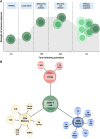
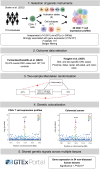
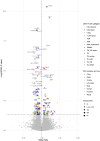
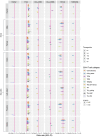
Recent research suggests higher circulating lymphocyte counts may protect against colorectal cancer (CRC). However, the role of specific lymphocyte subtypes and activation states remain unclear. CD4+ T cells-a highly dynamic lymphocyte subtype-undergo gene expression changes upon activation that are critical to their effector function. Previous studies using bulk tissue have limited our understanding of their role in CRC risk to static associations. We applied Mendelian randomization (MR) and genetic colocalisation to evaluate causal relationships of gene expression on CRC risk across multiple CD4+ T cell subtypes and activation states. Genetic proxies were obtained from single-cell transcriptomic data, allowing us to investigate the causal effect of expression of 1,805 genes across CD4+ T cell activation states on CRC risk (78,473 cases; 107,143 controls). Analyses were stratified by CRC anatomical subsites and sex, with sensitivity analyses assessing whether the observed effect estimates were likely to be CD4+ T cell-specific. We identified 6 genes-FADS2, FHL3, HLA-DRB1, HLA-DRB5, RPL28, and TMEM258-with strong evidence for a causal role in CRC development (FDR-P < 0.05; colocalisation H4 > 0.8). Causal estimates varied by CD4+ T cell subtype, activation state, CRC subsite and sex. However, many of genetic proxies used to instrument gene expression in CD4+ T cells also act as eQTLs in other tissues, highlighting the challenges of using genetic proxies to instrument tissue-specific expression changes. We demonstrate the importance of capturing the dynamic nature of CD4+ T cells in understanding CRC risk, and prioritize genes for further investigation in cancer prevention.
Keywords: CD4+ T cells; Mendelian randomization; colorectal cancer; gene expression; genetic epidemiology.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Leukocyte Biology.
Figures

Update of
-
Transcriptome-wide Mendelian randomisation exploring dynamic CD4+ T cell gene expression in colorectal cancer development.medRxiv [Preprint]. 2025 Apr 17:2025.04.15.25325863. doi: 10.1101/2025.04.15.25325863. medRxiv. 2025. Update in: J Leukoc Biol. 2025 Oct 1;117(10):qiaf131. doi: 10.1093/jleuko/qiaf131. PMID: 40321251 Free PMC article. Updated. Preprint.
References
-
- World Health Organization . Colorectal cancer; 2023. https://www.who.int/news-room/fact-sheets/detail/colorectal-cancer
-
- World Cancer Research Fund, American Institute for Cancer Research . Diet, Nutrition, Physical Activity and Colorectal Cancer: a Global Perspective. Continuous Update Project Expert Report 2018; 2018. dietandcancerreport.org
MeSH terms
Grants and funding
- AZV NU22J-03-00033/Ministry of Health of the Czech Republic
- U01 CA167551/NH/NIH HHS/United States
- C18281/A29019/Cancer Research UK 25
- C18281/A29019/CRUK Integrative Cancer Epidemiology Programme
- U19 CA148107/GF/NIH HHS/United States
- C18281/A30905/Cancer Research UK Population Research Committee
- HHSN268201200008C/HL/NHLBI NIH HHS/United States
- R01 CA081488/CA/NCI NIH HHS/United States
- HHSN268201200008I/GF/NIH HHS/United States
- U01 CA122839/CA/NCI NIH HHS/United States
- 218495/Z/19/Z/WT_/Wellcome Trust/United Kingdom
- U01 CA122839/NH/NIH HHS/United States
- MC_UU_00032/03/MRC_/Medical Research Council/United Kingdom
- U19 CA148107/CA/NCI NIH HHS/United States
- R01 CA81488/GF/NIH HHS/United States
- U01 CA167551/CA/NCI NIH HHS/United States
- R01 CA143247/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- University of Bristol
- LX22NPO5102/National Institute for Cancer Research
- HHSN268201200008I/HL/NHLBI NIH HHS/United States
- 001/WHO_/World Health Organization/International
LinkOut - more resources
Full Text Sources
Medical
Research Materials

