Combined effects of ciprofloxacin and microplastics on alpine spring water microbiota: evidence from glacier-fed microcosm experiments
- PMID: 40980327
- PMCID: PMC12443835
- DOI: 10.3389/fmicb.2025.1654589
Combined effects of ciprofloxacin and microplastics on alpine spring water microbiota: evidence from glacier-fed microcosm experiments
Abstract
Introduction: Emerging contaminants such as microplastics (MPs) and antibiotics pose increasing environmental and public health risks due to their persistence and incomplete removal by wastewater treatment processes. MPs can act as vectors for antibiotics, facilitating their environmental spreading and supporting biofilm formation, which can enhance horizontal gene transfer and antibiotic resistance. This study investigates the combined effects of ciprofloxacin (CIP) and polyethylene terephthalate (PET) MPs on microbiota in alpine spring water (SW) sourced from a rock glacier.
Methods: Four experimental scenarios (Control, CIP, PET, CIP + PET) were established to assess the sorption dynamics of CIP onto PET particles and the consequent microbial responses. A multidisciplinary analytical approach combining ultra-performance liquid chromatography, microscopy, quantitative PCR, and metabarcoding was applied.
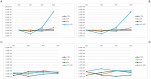
Results: CIP exhibited progressive sorption onto PET, accompanied by a time-dependent increase in biofilm formation, most pronounced in the CIP + PET condition. qPCR revealed elevated copy numbers of resistance genes qnrA and qnrB in CIP + PET, suggesting synergistic effects between antibiotics and MPs in promoting resistance. CIP was the dominant driver of microbial compositional shifts, favoring known CIP-degrading taxa. A shared core microbiome of 216 amplicon sequence variants was detected across all conditions, but specific taxa were differentially enriched under varying exposures. The combined CIP + PET test induced the strongest community shifts, while CIP alone shared fewer taxa with controls, indicating selective pressure for resistant microorganisms like Achromobacter. PET MPs also shaped distinct microbial assemblages, possibly by offering niches favoring biofilm-associated genera such as Luteolibacter. Biodiversity metrics showed highest richness and evenness in CIP-free conditions (Control and PET), while CIP significantly reduced alpha diversity, favoring resistant taxa, as confirmed by NMDS and lower Shannon and Simpson indices. Effects of MPs were still noticeable.
Conclusion: These findings demonstrate the disruptive effects of CIP on alpine freshwater microbial communities and highlight the additional, though more moderate, influence of MPs. The combined presence of MPs and antibiotics may exacerbate resistance spreading by enhancing persistence and providing favorable conditions for resistant biofilms. A mechanistic understanding of these interactions is essential for accurate risk assessment and the development of effective mitigation strategies in alpine and other vulnerable freshwater ecosystems.
Keywords: alpine ecosystem; antibiotic resistance; emerging contaminants; freshwater; microplastics pollution; polyethylene terephthalate.
Copyright © 2025 Mosca Angelucci, Piergiacomo, Donati, Pagani, Minuti, Brusetti and Tomei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Bach E., Radić V., Schoof C. (2018). How sensitive are mountain glaciers to climate change? Insights from a block model. J. Glaciol. 64, 247–258. doi: 10.1017/jog.2018.15 - DOI
-
- Briales A., Rodríguez-Martínez J. M., Velasco C., de Alba P. D., Martínez-Martínez L., Pascual A. (2012). SOS response induction by levofloxacin in qnrB1 and qnrD1 producing Escherichia coli. J. Antimicrob. Chemother. 67, 2676–2679. doi: 10.1093/jac/dks326 - DOI
-
- Buta M., Hubeny J., Zieliński W., Harnisz M., Korzeniewska E. (2021). Sewage sludge in agriculture: The effects of selected chemical pollutants and emerging genetic resistance determinants on the quality of soil and crops – A review. Ecotoxicol. Environ. Saf. 214:112070. doi: 10.1016/j.ecoenv.2021.112070, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

