Temperature-driven biogeography of marine giant viruses infecting picoeukaryotes Micromonas
- PMID: 40980734
- PMCID: PMC12448749
- DOI: 10.1093/ismeco/ycaf137
Temperature-driven biogeography of marine giant viruses infecting picoeukaryotes Micromonas
Abstract
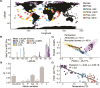
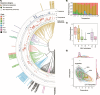
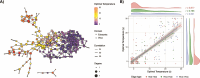
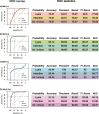
Climate shapes the biogeography of microbial and viral communities in the ocean. Among abiotic factors, temperature is one of the main drivers of microbial community distribution. However, we lack knowledge on how temperature shapes the life history traits, population dynamics, and the biogeography of marine viruses. This study integrates mathematical modeling with in situ observations to investigate the temperature-driven biogeography of marine viruses. We focused on prasinoviruses, a group of giant viruses that infect the picoeukaryote Micromonas, a widespread phytoplankton with thermotypes adapted from poles to tropics. Analyzing the Tara Oceans and Polar Circle databases, we found that temperature is the primary determinant of Micromonas virus (MicV) distribution in the surface ocean. Phylogenetic reconstruction of MicVs revealed that these viruses form several groups with cryophile or cryo-mesophile preferences. We applied a mechanistic model to describe temperature-driven population dynamics, allowing us to predict the global presence and absence of MicVs. The probability of lysis and the probability of infection emerged as reliable predictors of MicV distribution, indicating that temperature-driven cellular mechanisms significantly shape viral community structure and distribution in the global oceans.
Keywords: Micromonas; TARA; biogeography; ecological modeling; marine viruses; phytoplankton; temperature.
© The Author(s) 2025. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures

References
LinkOut - more resources
Full Text Sources

