Comparative single-cell and spatial profiling of anti-SSA-positive and anti-centromere-positive Sjögren's disease reveals common and distinct immune activation and fibroblast-mediated inflammation
- PMID: 40983612
- PMCID: PMC12454658
- DOI: 10.1038/s41467-025-63935-9
Comparative single-cell and spatial profiling of anti-SSA-positive and anti-centromere-positive Sjögren's disease reveals common and distinct immune activation and fibroblast-mediated inflammation
Abstract
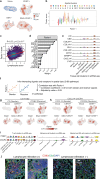
Sjögren's disease (SjD) is an autoimmune disease that causes salivary gland dysfunction due to immune-mediated destruction. While autoantibodies such as anti-SSA and anti-centromere (CENT) are associated with distinct clinical manifestations, the molecular features remain to be elucidated. In this study, we apply multi-modal single-cell technologies: single-cell RNA sequencing, T cell and B cell receptor sequencing and spatial transcriptomics to salivary gland lesions, aiming to elucidate common and unique cellular and transcriptional signatures linked to different autoantibody profiles. Our analysis demonstrates that GZMB+GNLY+ CD8+ T cells are the main expanded subset across different autoantibody statuses, highlighting their central role in SjD pathogenesis, while the enrichment of memory B cells is more prominent in anti-CENT-positive patients. Cytokine signaling also differs by autoantibody profile, with an activated interferon signature in anti-SSA-positive patients, whereas TGFβ signaling is enhanced in anti-CENT-positive patients. Furthermore, spatial profiling reveals THY1+ fibroblasts, expressing complement genes and chemokines, as key hubs orchestrating inflammation within the salivary glands. These findings deepen our understanding of the pathogenesis of SjD, and may inform the development of targeted and personalized therapeutic strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Mariette, X. & Criswell, L. A. Primary Sjögren’s syndrome. N. Engl. J. Med.378, 931–939 (2018). - PubMed
-
- Beydon, M. et al. Epidemiology of Sjögren syndrome. Nat. Rev. Rheumatol.20, 158–169 (2023). - PubMed
-
- Nocturne, G. & Mariette, X. Advances in understanding the pathogenesis of primary Sjögren’s syndrome. Nat. Rev. Rheumatol.9, 544–556 (2013). - PubMed
-
- Lin, X. et al. Th17 cells play a critical role in the development of experimental Sjögren’s syndrome. Ann. Rheum. Dis.74, 1302–1310 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

