NR3C1-mediated epigenetic regulation suppresses astrocytic immune responses in mice
- PMID: 40983615
- PMCID: PMC12454645
- DOI: 10.1038/s41467-025-64088-5
NR3C1-mediated epigenetic regulation suppresses astrocytic immune responses in mice
Abstract
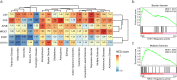
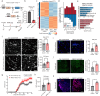
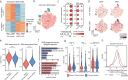
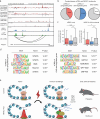
Astrocytes are critical contributors to brain disorders, yet the mechanisms underlying their selective vulnerability to specific diseases remain poorly understood. Here, we demonstrate that NR3C1 acts as a key regulator of early postnatal astrocyte development, shaping long-term immune responses in mice. Through integrative analyses of gene expression, chromatin accessibility, and long-range chromatin interactions, we identify 55 stage-specific TFs, with NR3C1 uniquely associated with early postnatal maturation. Although mice lacking astrocytic NR3C1 exhibit no detectable developmental abnormalities, these mice display heightened susceptibility to exacerbated immune responses following adult-onset experimental autoimmune encephalomyelitis (EAE). Many of the dysregulated EAE response genes are linked to candidate cis-regulatory elements altered by early NR3C1 loss, driving exacerbated inflammatory responses. Notably, only NR3C1 depletion during early, but not late, astrocyte development induces long-lasting epigenetic reprogramming that primes astrocytic immune responses.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Kenny, E. M. et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol. Psychiatry19, 872–879 (2014). - PubMed
-
- Knott, C., Wilkin, G. P. & Stern, G. Astrocytes and microglia in the substantia nigra and caudate-putamen in Parkinson’s disease. Parkinsonism Relat. Disord.5, 115–122 (1999). - PubMed
MeSH terms
Substances
Grants and funding
- 2023R1A2C3002773/National Research Foundation of Korea (NRF)
- 2022R1A5A1026413/National Research Foundation of Korea (NRF)
- RS-2023-00262527/National Research Foundation of Korea (NRF)
- RS-2023-00223069/National Research Foundation of Korea (NRF)
- 2021R1A2C3005704/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources

