The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases
- PMID: 40983680
- PMCID: PMC12552125
- DOI: 10.1038/s42255-025-01365-z
The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases
Abstract
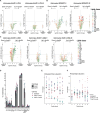
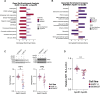
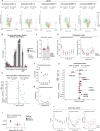
Lipid alterations in the brain have been implicated in many neurodegenerative diseases. To facilitate comparative lipidomic research across brain diseases, we establish a data common named the Neurolipid Atlas that we prepopulated with isogenic induced pluripotent stem cell (iPS cell)-derived lipidomics data for different brain diseases. Additionally, the resource contains lipidomics data of human and mouse brain tissue. Leveraging multiple datasets, we demonstrate that iPS cell-derived neurons, microglia and astrocytes exhibit distinct lipid profiles that recapitulate in vivo lipotypes. Notably, the Alzheimer disease (AD) risk gene ApoE4 drives cholesterol ester (CE) accumulation specifically in human astrocytes and we also observe CE accumulation in whole-brain lipidomics from persons with AD. Multiomics interrogation of iPS cell-derived astrocytes revealed that altered cholesterol metabolism has a major role in astrocyte immune pathways such as the immunoproteasome and major histocompatibility complex class I antigen presentation. Our data commons, available online ( https://neurolipidatlas.com/ ), allows for data deposition by the community and provides a user-friendly tool and knowledge base for a better understanding of lipid dyshomeostasis in neurodegenerative diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.K. has acted as a consultant for NewAmsterdam Pharma. M.K. is a coscientific founder of Montara Therapeutics, serves on the scientific advisory boards of Engine Biosciences, Casma Therapeutics, Cajal Neuroscience, Alector and Montara Therapeutics and is an advisor to Modulo Bio and Recursion Therapeutics. M.G. is a consultant to Boehringer-Ingelheim Pharma and an inventor of patent application WO2023172132A1. The other authors declare no competing interests.
Figures










Update of
-
The Neurolipid Atlas: a lipidomics resource for neurodegenerative diseases uncovers cholesterol as a regulator of astrocyte reactivity impaired by ApoE4.bioRxiv [Preprint]. 2024 Jul 3:2024.07.01.601474. doi: 10.1101/2024.07.01.601474. bioRxiv. 2024. Update in: Nat Metab. 2025 Oct;7(10):2142-2164. doi: 10.1038/s42255-025-01365-z. PMID: 39005258 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- 733050515/ZonMw (Netherlands Organisation for Health Research and Development)
- WE-03-2017-09P/Alzheimer Nederland (Alzheimer Netherlands)
- ADSF-21-831212-C/ALZ/Alzheimer's Association/United States
- T32NS115706/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- T32 NS115706/NS/NINDS NIH HHS/United States
- 2019-AARG-643165/ALZ/Alzheimer's Association/United States
- P01 NS084974-06A1/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- 435005033/ZonMw (Netherlands Organisation for Health Research and Development)
- P30 CA062203/CA/NCI NIH HHS/United States
- WE.03-2019-13/Alzheimer Nederland (Alzheimer Netherlands)
- ADSF-21-816750/ALZ/Alzheimer's Association/United States
- R01 AG062359/AG/NIA NIH HHS/United States
- R35 NS116872/NS/NINDS NIH HHS/United States
- U01 AG072464/AG/NIA NIH HHS/United States
- WE.15-2019-16/Alzheimer Nederland (Alzheimer Netherlands)
- P01 NS084974/NS/NINDS NIH HHS/United States
- 09150172110086/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- EDUC412812/California Institute for Regenerative Medicine (CIRM)
LinkOut - more resources
Full Text Sources
Medical

