MHCquant2 refines immunopeptidomics tumor antigen discovery
- PMID: 40983925
- PMCID: PMC12455830
- DOI: 10.1186/s13059-025-03763-8
MHCquant2 refines immunopeptidomics tumor antigen discovery
Abstract
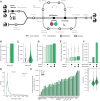
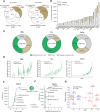
Confident identification of human leukocyte antigen (HLA)-presented peptides is crucial for advancing cancer immunotherapy. We present MHCquant2, a scalable and modular Nextflow pipeline integrated into nf-core as a reproducible, portable, and standardized workflow for immunopeptidomics. This integration allows a community-driven, robust solution for high-throughput analyses across operating systems and cloud infrastructures. MHCquant2 integrates open-source tools including OpenMS, DeepLC, and MS2PIP, improving peptide identifications by up to 27% across diverse MS platforms, particularly enriching low-abundant peptides. MHCquant2 demonstrates state-of-the-art performance on our novel benignMHCquant2 dataset (n = 92) and expands the benign human immunopeptidome by over 160,000 unique naturally presented HLA peptides.
Keywords: HLA; Immunopeptidomics; Immunotherapy; Mass spectrometry; Nextflow; Nf-core; Open-source; Pipeline.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Cantonal Ethics Committee Zürich (KEK) (BASEC-Nr. Req-2016–00604). For none of the included patients, a refusal of post-mortem contribution to medical research was documented, and study procedures are in accordance with applicable Swiss law for research on humans (Bundesgesetz über die Forschung am Menschen, Art. 38). In addition, the study protocol was reviewed by the ethics committee at the University of Tübingen and received a favorable assessment without any objections to the study conduct (Project Nr. 364/2017BO2). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Hilf N, Kuttruff-Coqui S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–5. - PubMed
-
- Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–6. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

