Hydrogel Loaded with Aminoethyl Anisamide-Modified Exosomes Attenuates Hepatic Fibrosis by Targeting Activated Hepatic Stellate Cells
- PMID: 40985972
- PMCID: PMC12509324
- DOI: 10.1021/acsnano.5c06003
Hydrogel Loaded with Aminoethyl Anisamide-Modified Exosomes Attenuates Hepatic Fibrosis by Targeting Activated Hepatic Stellate Cells
Abstract
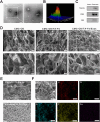
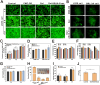
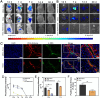
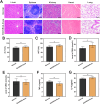
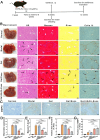
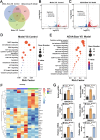
Stem cell-based regenerative research has highlighted the therapeutic potential of human umbilical cord mesenchymal stem cell-derived exosomes (hucMSC-Exos) for hepatic tissue regeneration and repair. However, exosomes undergo rapid clearance following systemic administration, limiting their therapeutic potential because of insufficient retention and sustained release. In this study, an innovative hydrogel-mediated delivery platform encapsulating aminoethyl anisamide (AEAA)-functionalized exosomes was developed to mitigate hepatic fibrosis. By synthesizing a hydrogel (CMC-OD/TA-Fe(III), Gel) composed of carboxymethyl chitosan, oxidized dextran, and iron tannate, and then encapsulating umbilical cord mesenchymal stem cell-derived exosomes functionalized by AEAA (AEAA-Exos), we implanted this Gel/AEAA-Exos into mice with hepatic fibrosis by intraperitoneal injection to evaluate the therapeutic effect of the hydrogel. The hydrogel had favorable physical properties, optimal biocompatibility, and a sustained-release profile. And Gel/AEAA-Exos system significantly reduced oxidative stress and alleviated hepatic fibrosis. Additionally, RNA-seq revealed that the Gel/AEAA-Exos system ameliorates hepatic fibrogenesis mainly by modulating oxidative stress, collagen deposition, and inflammatory cascade in liver tissues. This strategy offers a targeted and efficient approach for treating liver fibrosis induced by chronic hepatic injury and improves targeting efficiency and therapeutic outcomes through engineered exosome delivery.
Keywords: AEAA-engineered exosomes; antihepatic fibrosis; hydrogel; sustained release; target.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

