Crossover designation recruits condensin to reorganize the meiotic chromosome axis
- PMID: 40987260
- PMCID: PMC12533695
- DOI: 10.1016/j.cub.2025.08.019
Crossover designation recruits condensin to reorganize the meiotic chromosome axis
Abstract
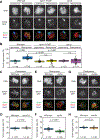
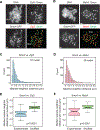
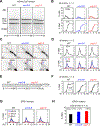
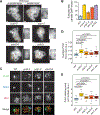
Crossover recombination supports meiotic chromosome inheritance and fertility by establishing chiasmata between homologous chromosomes prior to the first meiotic division. In addition to the physical exchange of DNA mediated by meiotic recombination, chiasma formation also involves restructuring of the underlying chromosome axis, possibly to help with chiasma maturation or to resolve chromosomal interlocks. Here, we identify condensin as an important regulator of axis remodeling in S. cerevisiae. Condensin is recruited near sites of meiotic crossover designation by pro-crossover factors but is largely dispensable for DNA exchange. Instead, condensin helps to create discontinuities in the meiotic chromosome axis by promoting removal of cohesin. In addition, chromosomes of condensin mutants exhibit unusually common parallel chromatin clouds and experience a chromosomal buildup of the conserved axis remodeler Pch2. Consistent with an important role of axis restructuring at crossover sites, the canonical anaphase-bridge phenotype of condensin mutants is partly rescued by redirecting meiotic DNA repair to sister chromatids instead of homologous chromosomes, suggesting that crossover-associated axis reorganization is important for faithful meiotic chromosome segregation.
Keywords: Pch2; Rec8; Red1; ZMM factors; axial element; synapsis.
Copyright © 2025 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Crossover designation recruits condensin to reorganize the meiotic chromosome axis.bioRxiv [Preprint]. 2025 Sep 16:2020.07.16.207068. doi: 10.1101/2020.07.16.207068. bioRxiv. 2025. Update in: Curr Biol. 2025 Sep 22;35(18):4537-4552.e6. doi: 10.1016/j.cub.2025.08.019. PMID: 40625642 Free PMC article. Updated. Preprint.
References
-
- Bickmore WA, and van Steensel B (2013). Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

