One-carbon metabolism modulates miR-29a-DNA methylation crosstalk in Alzheimer's disease
- PMID: 40987757
- PMCID: PMC12457075
- DOI: 10.1002/alz.70703
One-carbon metabolism modulates miR-29a-DNA methylation crosstalk in Alzheimer's disease
Abstract
Introduction: Alzheimer's disease (AD)'s multifactorial nature stresses the role of epigenetics in affecting different pathological pathways. We demonstrated that one-carbon metabolism epigenetically impacts AD-like phenotype. Here, we investigated the crosstalk between methylation and microRNAs in AD.
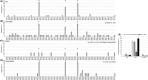
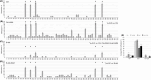
Methods: We altered one-carbon metabolism to induce hypo- and hyper-methylation, in SK-N-BE neuroblastoma cells and TgCRND8 mice. miRNAs were profiled through a polymerase chain reaction array, then we focused on miR-29a expression and methylation of its genomic locus. Finally, we assessed miR-29a expression and methylation in the brain of AD subjects.
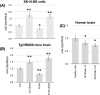
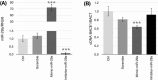
Results: MiR-29a was repressed in hypomethylating and expressed in hypermethylating conditions. The expression of miR-29a and of its target, BACE1, was inversely correlated.
Discussion: We demonstrated for the first time that miR-29a is modulated by one-carbon metabolism through DNA methylation, disclosing the molecular mechanisms regulating BACE1 expression in AD. These data confirm miR-29a's protective role in AD and support miR-29a as a potential biomarker for AD.
Keywords: Alzheimer's disease; DNA methylation; epigenetics; microRNAs; mir‐29a; non‐CPG methylation; one‐carbon metabolism.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Tiziana Raia, Luiza Diniz Ferreira Borges, Rosaria A. Cavallaro, Isidre Ferrer, Stefano Cinti, Mariano Bizzarri, and Marco Lucarelli have nothing to disclose. Andrea Fuso is a consultant at “Gnosis by Lesaffre.” Author disclosures are available in the supporting information
Figures
References
-
- Parker J, Moris JM, Goodman LC, et al. A multifactorial lens on risk factors promoting the progression of Alzheimer's disease. Brain Res. 2024;1846:149262. - PubMed
-
- Bocti C. Is simpler always better? The case for going beyond the amyloid hypothesis in Alzheimer's disease. J Alzheimers Dis. 2024;100(3):787‐789. - PubMed
-
- Castellani RJ, Jamshidi P, Plascencia‐Villa G, Perry G. The amyloid cascade hypothesis: a conclusion in search of support. Am J Pathol. 2024;S0002‐9440(24):00407‐00403. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

