Evaluation of HIV-1 DNA resistance evolution in highly treatment-experienced and multi-resistant individuals under suppressive antiretroviral therapy: a longitudinal study from the PRESTIGIO Registry
- PMID: 40990139
- PMCID: PMC12596042
- DOI: 10.1093/jac/dkaf349
Evaluation of HIV-1 DNA resistance evolution in highly treatment-experienced and multi-resistant individuals under suppressive antiretroviral therapy: a longitudinal study from the PRESTIGIO Registry
Abstract
Background: This study aimed to clarify whether resistance detected in HIV-1 DNA might evolve in virologically suppressed highly treatment-experienced (HTE) individuals with multidrug resistance (MDR).
Methods: Twenty-three virologically suppressed HTE MDR individuals from the PRESTIGIO Registry with two longitudinal samples available under virological suppression at two different time points (T0-T1) were analysed. HIV-1 DNA levels were quantified using droplet digital PCR, and resistance was assessed through next-generation sequencing (NGS) set at 5%. Mutational load was also evaluated.
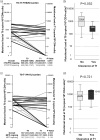
Results: At T0, individuals had been virologically suppressed for a median time of 3 years (IQR 3-5) under a salvage regimen, mostly containing dolutegravir (95.7%) and/or darunavir (69.6%). The median HIV-1 DNA level was 2588 copies/106 CD4+ cells at T0 and remained stable at T1 (2322 copies/106 CD4+ cells; P = 0.831). Individuals with at least ≥3-class resistance in HIV-1 DNA were 20 (87.0%) at T0 and 18 (78.2%) at T1 (P = 0.607). In those receiving NNRTI-sparing treatment (52.2%), the number of NNRTI major resistance mutation (MRM) significantly decreased over time (T0, 2 [1-3]; T1, 0 [0-1]; P = 0.027). No significant temporal differences in the number of PI, NRTI and integrase strand transfer inhibitor (INSTI) MRM were found. Specific MRM, such as M184V, decreased over time, particularly in individuals who were receiving a 3TC-/FTC-sparing salvage regimen or with a T0 mutational load of <1000 copies/106 CD4+ cells.
Conclusions: Over a year, HIV-1 DNA MRM generally remained unchanged in suppressed HTE MDR people with HIV (PWH) except for a significant decline in M184V and a reduction of NNRTI resistance in the absence of NNRTI pressure.
© The Author(s) 2025. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures

References
-
- Armenia D, Spagnuolo V, Bellocchi MC et al. Use of next-generation sequencing on HIV-1 DNA to assess archived resistance in highly treatment-experienced people with multidrug-resistant HIV under virological control: data from the PRESTIGIO Registry. J Antimicrob Chemother 2024; 79: 2354–63. 10.1093/jac/dkae236 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

