Accessible, uniform protein property prediction with a scikit-learn based toolset AIDE
- PMID: 40991335
- PMCID: PMC12553329
- DOI: 10.1093/bioinformatics/btaf544
Accessible, uniform protein property prediction with a scikit-learn based toolset AIDE
Abstract
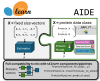
Summary: Protein property prediction via machine learning with and without labeled data is becoming increasingly powerful, yet methods are disparate and capabilities vary widely over applications. The software presented here, "Artificial Intelligence Driven protein Estimation (AIDE)", enables instantiating, optimizing, and testing many zero-shot and supervised property prediction methods for variants and variable length homologs in a single, reproducible notebook or script by defining a modular, standardized application programming interface (API), i.e. drop-in compatible with scikit-learn transformers and pipelines.
Availability and implementation: AIDE is an installable, importable python package inheriting from scikit-learn classes and API and is installable on Windows, Mac, and Linux. Many of the wrapped models internal to AIDE will be effectively inaccessible without a GPU, and some assume CUDA. The newest stable, tested version can be found at https://github.com/beckham-lab/aide_predict and a full user guide and API reference can be found at https://beckham-lab.github.io/aide_predict/. Static versions of both at the time of writing can be found on Zenodo.
© The Author(s) 2025. Published by Oxford University Press.
Figures

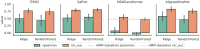
References
-
- Bank C. Epistasis and adaptation on fitness landscapes. Annu Rev Ecol Evol Syst 2022;53:457–79.
-
- Block P, Paern J, Hüllermeier E et al. Physicochemical descriptors to discriminate protein–protein interactions in permanent and transient complexes selected by means of machine learning algorithms. Proteins Struct Funct Bioinf 2006;65:607–22. - PubMed
-
- Bourque P, Dupuis R, Abran A et al. Fundamental principles of software engineering—a journey. J Syst Softw 2002;62:59–70.
MeSH terms
Substances
Grants and funding
- DE-AC36-08GO28308/National Renewable Energy Laboratory for the US Department of Energy (DOE)
- DE-SC0023278/US Department of Energy Office of Science Biological and Environmental Research
- US Department of Energy Office of Energy Efficiency and Renewable Energy Bioenergy Technologies Office (BETO)
- Agile BioFoundry
- DE-SC0022024/U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research
LinkOut - more resources
Full Text Sources
Miscellaneous

