DNA demethylation mediated endogenous retroviruses transcription promotes aberrant T cell differentiation in systemic lupus erythematosus via RIG-I pathway
- PMID: 40992212
- PMCID: PMC12492037
- DOI: 10.1016/j.ebiom.2025.105934
DNA demethylation mediated endogenous retroviruses transcription promotes aberrant T cell differentiation in systemic lupus erythematosus via RIG-I pathway
Abstract
Background: Systemic lupus erythematosus (SLE) displays quantitative and/or qualitative deficiencies of regulatory T cells (Treg) and expansion and hyperfunction of pathogenic T cells. However, the underlying mechanism of dysregulated T lymphocyte differentiation in SLE remains unclear.
Methods: Transcriptome sequencing and functional assays were performed to elucidate the mechanisms and function of human endogenous retroviruses (HERVs) on T cell differentiation in SLE. The effect of retinoic acid-inducible gene I (RIG-I) deficiency on lupus pathogenesis were assessed in lupus-like mouse models.
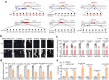
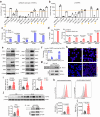
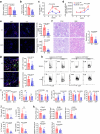
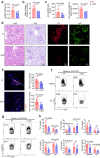
Findings: We found that many transcripts derived from HERVs were highly expressed in CD4+ T cells from patients with SLE due to DNA hypomethylation, some of which were characterized by double strand RNAs (dsRNAs). Excessive dsRNAs promoted Th1 cell differentiation and inhibited Treg cell differentiation via the activation of the dsRNA sensor RIG-I pathway, accompanied by a high level of type I interferons (IFN-I) and interferon-stimulated gene expression. In contrast, T cell-specific ablation of RIG-I alleviated disease progression in lupus-like mouse models by reducing the proportion of pathogenic T cells, restoring the percentage of Treg cells, and diminishing cytokine and autoantibody release. Importantly, we demonstrated that the dsRNA-induced RIG-I/IRF3 pathway regulated Th1 cell differentiation in a IFN-I/STAT1 signalling-dependent manner and inhibited Treg cell differentiation via SMAD3 signalling.
Interpretation: Our findings reveal the roles of HERV-derived dsRNA/RIG-I pathway in regulating the aberrant differentiation of T cells in patients with SLE and may facilitate the development of potential therapeutic targets for SLE.
Funding: A full list of funding sources can be found in the Funding section.
Keywords: Human endogenous retroviruses; RIG-I pathway; Systemic lupus erythematosus; T cell differentiation.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing financial interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

