CD103-CD8+ T cells promote neurotoxic inflammation in Alzheimer's disease via granzyme K-PAR-1 signaling
- PMID: 40993111
- PMCID: PMC12460627
- DOI: 10.1038/s41467-025-62405-6
CD103-CD8+ T cells promote neurotoxic inflammation in Alzheimer's disease via granzyme K-PAR-1 signaling
Abstract
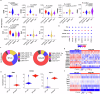
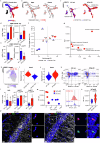
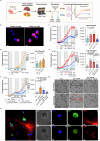
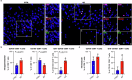
Immune mechanisms contribute to the neuropathology of Alzheimer's disease (AD) but the role of adaptive immune cells is unclear. Here we show that the brain CD8+ T cell compartment is dysregulated in AD patients and in the 3xTg-AD mouse model, accumulating activated CD103- tissue-resident memory T cells that produce large amounts of granzyme K (GrK). These CD103-CD8+ T cells originate from the circulation and migrate into the brain using LFA-1 integrin. Ablation of brain CD103-CD8+ T cells in 3xTg-AD mice ameliorates cognitive decline and reduces neuropathology. GrK induces neuronal dysfunction and tau hyperphosphorylation in human and mouse cells via protease-activated receptor-1 (PAR-1), which is expressed at higher levels in the AD brain, revealing a key immune-mediated neurotoxic axis. We conclude that communication between CD8+ T cells and the nervous system is altered in AD, paving the way for therapies targeting T cell-dependent neurotoxic inflammation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- 695714 Immunoalzheimer/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 101069397 NeutrAD/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 693606 IMPEDE/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

